7HVA
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0032363-001 (A71EV2A-x3480) | | Descriptor: | (3S)-N~1~-[3-(cyanomethyl)phenyl]piperidine-1,3-dicarboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7HVB
 
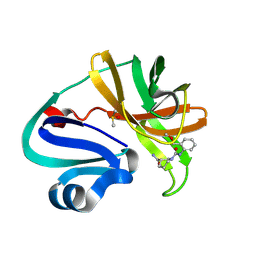 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0032369-001 (A71EV2A-x3483) | | Descriptor: | (3S)-N~1~-[2-(methoxymethyl)phenyl]piperidine-1,3-dicarboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.461 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7HVC
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0032359-001 (A71EV2A-x3485) | | Descriptor: | (3S)-N~1~-(3-methoxyphenyl)piperidine-1,3-dicarboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.355 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7HVD
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0032330-001 (A71EV2A-x3488) | | Descriptor: | 3-[3-chloro-2-(2-methoxyacetamido)-5-methylphenoxy]benzamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.523 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7HVE
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0032371-001 (A71EV2A-x3489) | | Descriptor: | (3S)-1-{[(4R)-2-methyl-5,6,7,8-tetrahydroimidazo[1,2-a]pyridin-3-yl]acetyl}piperidine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.617 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7HVF
 
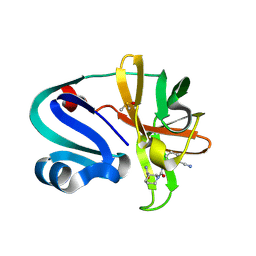 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0032365-001 (A71EV2A-x3509) | | Descriptor: | (3S)-N~1~-(3-cyanophenyl)piperidine-1,3-dicarboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7HVG
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0032380-001 (A71EV2A-x3510) | | Descriptor: | (3S)-N~1~-(2-methylphenyl)piperidine-1,3-dicarboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7HVH
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0032626-001 (A71EV2A-x3546) | | Descriptor: | (3S)-1-[(5-fluoro-2-methoxyphenyl)acetyl]piperidine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.608 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7HVI
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0032594-001 (A71EV2A-x3548) | | Descriptor: | (3S)-1-[(4-methoxypyridin-3-yl)acetyl]piperidine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7HVJ
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0032637-001 (A71EV2A-x3550) | | Descriptor: | (3S,6R)-1-[(2-methoxyphenyl)acetyl]-6-methylpiperidine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.339 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7HVK
 
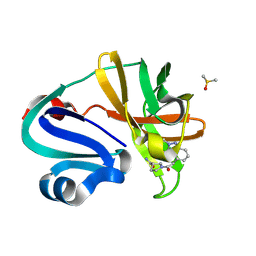 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0032647-001 (A71EV2A-x3556) | | Descriptor: | (3S)-1-[(2-methyl-2H-indazol-4-yl)acetyl]pyrrolidine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.734 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7HVL
 
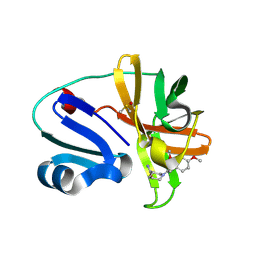 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0032685-001 (A71EV2A-x3557) | | Descriptor: | (3S)-1-[(2,4-dimethoxyphenyl)acetyl]piperidine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7HVM
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0032584-001 (A71EV2A-x3564) | | Descriptor: | DIMETHYL SULFOXIDE, N~3~-(2-methylphenyl)pyridine-3,5-dicarboxamide, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.378 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7HVN
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0032666-001 (A71EV2A-x3568) | | Descriptor: | 6-methyl-N-[2-(1,3-oxazol-4-yl)ethyl]-1H-indole-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7HVO
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0032676-001 (A71EV2A-x3571) | | Descriptor: | (3S)-1-[(2-methoxyphenyl)acetyl]azepane-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.361 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7HVQ
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0032627-001 (A71EV2A-x3575) | | Descriptor: | (3S)-1-[(3-cyano-2-methoxyphenyl)acetyl]piperidine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.455 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7HVR
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0032526-001 (A71EV2A-x3579) | | Descriptor: | (3S)-1-[(2-methoxyphenyl)methyl]-6-oxopiperidine-3-carboxamide, Protease 2A, ZINC ION | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.558 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7HVS
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0032461-001 (A71EV2A-x3581) | | Descriptor: | (3S)-1-[(2-methoxyphenyl)acetyl]pyrrolidine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7HVT
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0032067-002 (A71EV2A-x3593) | | Descriptor: | 1-methyl-N-[(2R)-1-(1,3-oxazol-4-yl)propan-2-yl]-1H-indazole-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7HVU
 
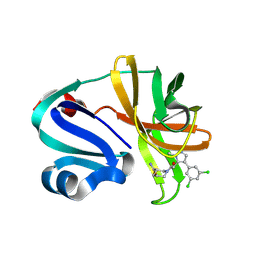 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0032503-001 (A71EV2A-x3606) | | Descriptor: | (3S)-1-{[4-(3,5-dichlorophenyl)-1-methyl-1H-pyrazol-5-yl]acetyl}piperidine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.429 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7HVV
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0032455-001 (A71EV2A-x3612) | | Descriptor: | (3S)-N~1~-(2-fluorophenyl)piperidine-1,3-dicarboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7HVW
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0032527-001 (A71EV2A-x3616) | | Descriptor: | (3S)-N~1~-[(2-methylphenyl)methyl]piperidine-1,3-dicarboxamide, Protease 2A, ZINC ION | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.339 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7HVX
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0032468-001 (A71EV2A-x3623) | | Descriptor: | (2S)-N-[(5-methyl-1,3-oxazol-2-yl)methyl]-2-phenylpropane-1-sulfonamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7HVY
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0032512-001 (A71EV2A-x3634) | | Descriptor: | (3S,4R)-1-[(2-methoxyphenyl)acetyl]-4-methylpyrrolidine-3-carboxamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7HVZ
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0032409-001 (A71EV2A-x3663) | | Descriptor: | (2S)-4-[(2-methoxyphenyl)acetyl]morpholine-2-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.208 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
