6ZSG
 
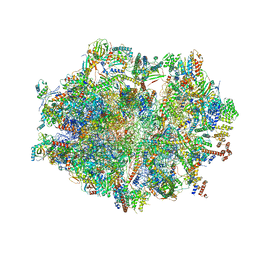 | | Human mitochondrial ribosome in complex with mRNA, A-site tRNA, P-site tRNA and E-site tRNA | | Descriptor: | 12S mitochondrial rRNA, 16S mitochondrial rRNA, 28S ribosomal protein S10, ... | | Authors: | Aibara, S, Singh, V, Modelska, A, Amunts, A. | | Deposit date: | 2020-07-15 | | Release date: | 2020-10-14 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of mitochondrial translation.
Elife, 9, 2020
|
|
6ZSP
 
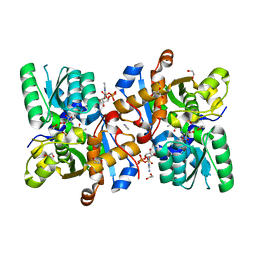 | | Human serine racemase bound to ATP and malonate. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Koulouris, C.R, Bax, B.D, Roe, S.M, Atack, J.R. | | Deposit date: | 2020-07-16 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Tyrosine 121 moves revealing a ligandable pocket that couples catalysis to ATP-binding in serine racemase.
Commun Biol, 5, 2022
|
|
6ZSQ
 
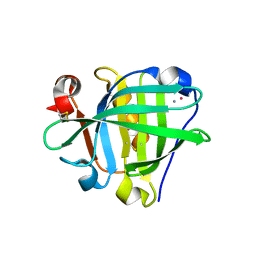 | | Crystal structure of the Cisplatin beta-Lactoglobulin adduct formed after 18 h of soaking | | Descriptor: | AMMONIA, Beta-lactoglobulin, PLATINUM (II) ION, ... | | Authors: | Balasco, N, Ferraro, G, Merlino, A. | | Deposit date: | 2020-07-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Cisplatin binding to beta-lactoglobulin: a structural study.
Dalton Trans, 49, 2020
|
|
6ZSR
 
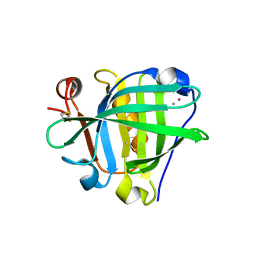 | | Crystal structure of the Cisplatin beta-Lactoglobulin adduct formed after 72 h of soaking | | Descriptor: | AMMONIA, Beta-lactoglobulin, PLATINUM (II) ION, ... | | Authors: | Balasco, N, Ferraro, G, Merlino, A. | | Deposit date: | 2020-07-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Cisplatin binding to beta-lactoglobulin: a structural study.
Dalton Trans, 49, 2020
|
|
6ZT0
 
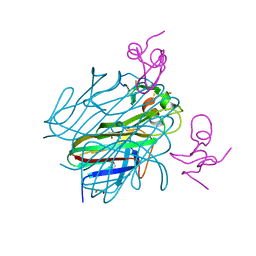 | | Crystal structure of the Eiger TNF domain/Grindelwald extracellular domain complex | | Descriptor: | 1,2-ETHANEDIOL, Protein eiger, Protein grindelwald | | Authors: | Palmerini, V, Cecatiello, V, Pasqualato, S, Mapelli, M. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Drosophila TNFRs Grindelwald and Wengen bind Eiger with different affinities and promote distinct cellular functions.
Nat Commun, 12, 2021
|
|
6ZTJ
 
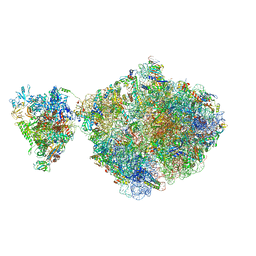 | | E. coli 70S-RNAP expressome complex in NusG-coupled state (38 nt intervening mRNA) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZTL
 
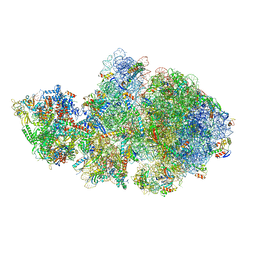 | | E. coli 70S-RNAP expressome complex in collided state bound to NusG | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZTM
 
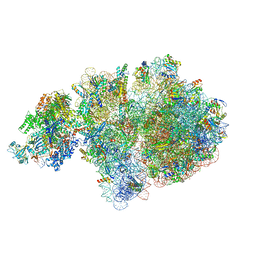 | | E. coli 70S-RNAP expressome complex in collided state without NusG | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZTN
 
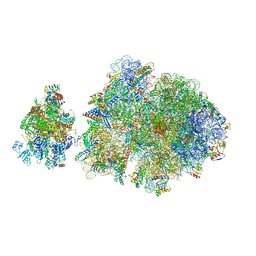 | | E. coli 70S-RNAP expressome complex in NusG-coupled state (42 nt intervening mRNA) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZTO
 
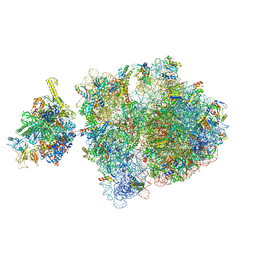 | | E. coli 70S-RNAP expressome complex in uncoupled state 1 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-23 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZTP
 
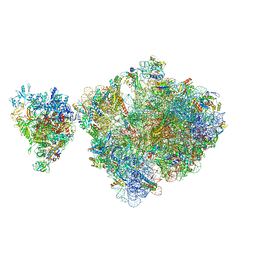 | | E. coli 70S-RNAP expressome complex in uncoupled state 6 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZU1
 
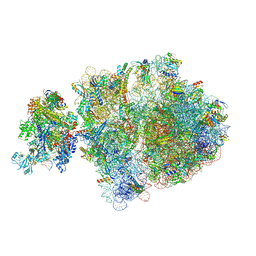 | | E. coli 70S-RNAP expressome complex in uncoupled state 2 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-21 | | Release date: | 2020-09-16 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZU2
 
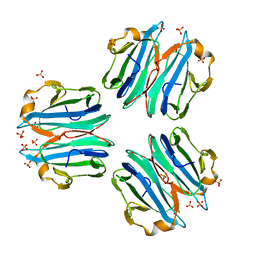 | | CML1 crystal structure in complex with H-type 1 trisaccharide | | Descriptor: | Mucin-binding lectin 1, SULFATE ION, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Varrot, A, Bleuler-Martinez, S. | | Deposit date: | 2020-07-21 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-function relationship of a novel fucoside-binding fruiting body lectin from Coprinopsis cinerea exhibiting nematotoxic activity.
Glycobiology, 32, 2022
|
|
6ZU5
 
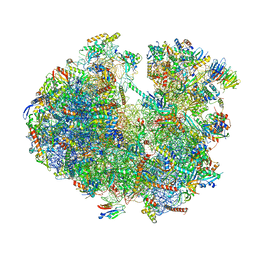 | | Structure of the Paranosema locustae ribosome in complex with Lso2 | | Descriptor: | 18S rRNA, 25S rRNA, 5S rRNA, ... | | Authors: | Ehrenbolger, K, Jespersen, N, Sharma, H, Sokolova, Y.Y, Tokarev, Y.S, Vossbrinck, C.R, Barandun, J. | | Deposit date: | 2020-07-21 | | Release date: | 2020-11-04 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Differences in structure and hibernation mechanism highlight diversification of the microsporidian ribosome.
Plos Biol., 18, 2020
|
|
6ZUJ
 
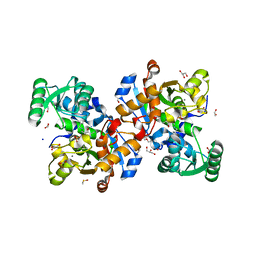 | | Human serine racemase holoenzyme from 20% DMSO soak (XChem crystallographic fragment screen). | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Koulouris, C.R, Bax, B.D, Roe, S.M, Atack, J.R. | | Deposit date: | 2020-07-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tyrosine 121 moves revealing a ligandable pocket that couples catalysis to ATP-binding in serine racemase.
Commun Biol, 5, 2022
|
|
6ZV5
 
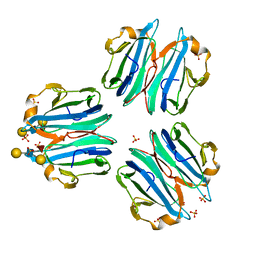 | | CML1 crystal structure in complex with Lewis a tetrasaccharide | | Descriptor: | Mucin-binding lectin 1, SULFATE ION, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Varrot, A, Bleuler-Martinez, S. | | Deposit date: | 2020-07-24 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-function relationship of a novel fucoside-binding fruiting body lectin from Coprinopsis cinerea exhibiting nematotoxic activity.
Glycobiology, 32, 2022
|
|
6ZVJ
 
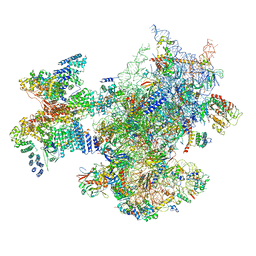 | | Structure of a human ABCE1-bound 43S pre-initiation complex - State II | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Kratzat, H, Mackens-Kiani, T, Ameismeier, A, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A structural inventory of native ribosomal ABCE1-43S pre-initiation complexes.
Embo J., 40, 2021
|
|
6ZVK
 
 | | The Halastavi arva virus (HalV) intergenic region IRES promotes translation by the simplest possible initiation mechanism | | Descriptor: | 18S RIBOSOMAL RNA, 28S RIBOSOMAL RNA, 40S RIBOSOMAL PROTEIN ES17, ... | | Authors: | Abaeva, I.S, Vicens, Q, Bochler, A, Soufari, H, Simonetti, A, Pestova, T, Hashem, Y, Hellen, C.U.T. | | Deposit date: | 2020-07-24 | | Release date: | 2020-12-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | The Halastavi arva Virus Intergenic Region IRES Promotes Translation by the Simplest Possible Initiation Mechanism.
Cell Rep, 33, 2020
|
|
6ZVM
 
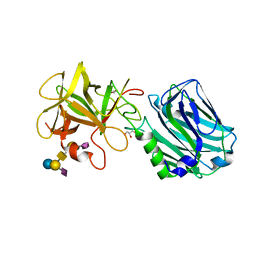 | | Botulinum neurotoxin B2 binding domain in complex with GD1a | | Descriptor: | GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Neurotoxin | | Authors: | Davies, J.R, Masuyer, G, Stenmark, P. | | Deposit date: | 2020-07-25 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biochemical Characterization of Botulinum Neurotoxin Subtype B2 Binding to Its Receptors.
Toxins, 12, 2020
|
|
6ZVN
 
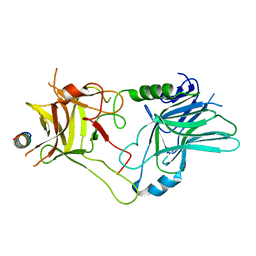 | |
6ZVR
 
 | | C11 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | Vipp1 | | Authors: | Liu, J.W, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZVS
 
 | | C12 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | Vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZVT
 
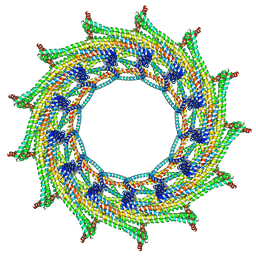 | | C13 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | Vipp1 | | Authors: | Liu, J.W, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZW4
 
 | | C14 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J.W, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZW5
 
 | | C15 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J.W, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|







