5WDT
 
 | | 70S ribosome-EF-Tu H84A complex with GppNHp | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fislage, M, Brown, Z, Frank, J. | | Deposit date: | 2017-07-06 | | Release date: | 2018-04-25 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM shows stages of initial codon selection on the ribosome by aa-tRNA in ternary complex with GTP and the GTPase-deficient EF-TuH84A.
Nucleic Acids Res., 46, 2018
|
|
5WE4
 
 | | 70S ribosome-EF-Tu wt complex with GppNHp | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fislage, M, Brown, Z, Frank, J. | | Deposit date: | 2017-07-07 | | Release date: | 2018-04-25 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM shows stages of initial codon selection on the ribosome by aa-tRNA in ternary complex with GTP and the GTPase-deficient EF-TuH84A.
Nucleic Acids Res., 46, 2018
|
|
5WE6
 
 | | 70S ribosome-EF-Tu H84A complex with GTP and cognate tRNA | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fislage, M, Frank, J. | | Deposit date: | 2017-07-07 | | Release date: | 2018-04-25 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM shows stages of initial codon selection on the ribosome by aa-tRNA in ternary complex with GTP and the GTPase-deficient EF-TuH84A.
Nucleic Acids Res., 46, 2018
|
|
5WF0
 
 | |
5WFK
 
 | |
5WFS
 
 | |
5WIS
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with methymycin and bound to mRNA and A-, P- and E-site tRNAs at 2.7A resolution | | Descriptor: | (3R,4S,5S,7R,9E,11S,12R)-12-ethyl-11-hydroxy-3,5,7,11-tetramethyl-2,8-dioxooxacyclododec-9-en-4-yl 3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranoside, 16S Ribosomal RNA, 23S ribosomal RNA, ... | | Authors: | Almutairi, M.M, Svetlov, M.S, Hansen, D.A, Khabibullina, N.F, Klepacki, D, Kang, H.Y, Sherman, D.H, Vazquez-Laslop, N, Polikanov, Y.S, Mankin, A.S. | | Deposit date: | 2017-07-20 | | Release date: | 2018-02-14 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Co-produced natural ketolides methymycin and pikromycin inhibit bacterial growth by preventing synthesis of a limited number of proteins.
Nucleic Acids Res., 45, 2017
|
|
5WIT
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with pikromycin and bound to mRNA and A-, P- and E-site tRNAs at 2.6A resolution | | Descriptor: | (3R,5R,6S,7S,9R,11E,13S,14R)-14-ethyl-13-hydroxy-3,5,7,9,13-pentamethyl-2,4,10-trioxo-1-oxacyclotetradec-11-en-6-yl 3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranoside, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Almutairi, M.M, Svetlov, M.S, Hansen, D.A, Khabibullina, N.F, Klepacki, D, Kang, H.Y, Sherman, D.H, Vazquez-Laslop, N, Polikanov, Y.S, Mankin, A.S. | | Deposit date: | 2017-07-20 | | Release date: | 2018-02-14 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Co-produced natural ketolides methymycin and pikromycin inhibit bacterial growth by preventing synthesis of a limited number of proteins.
Nucleic Acids Res., 45, 2017
|
|
5WJU
 
 | | Cryo-EM structure of B. subtilis flagellar filaments A39V, N133H | | Descriptor: | Flagellin | | Authors: | Wang, F, Burrage, A.M, Kearns, D.B, Egelman, E.H. | | Deposit date: | 2017-07-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | A structural model of flagellar filament switching across multiple bacterial species.
Nat Commun, 8, 2017
|
|
5WJV
 
 | |
5WJW
 
 | | Cryo-EM structure of B. subtilis flagellar filaments H84R | | Descriptor: | Flagellin | | Authors: | Wang, F, Burrage, A.M, Orlova, A, Kearns, D.B, Egelman, E.H. | | Deposit date: | 2017-07-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | A structural model of flagellar filament switching across multiple bacterial species.
Nat Commun, 8, 2017
|
|
5WJX
 
 | | Cryo-EM structure of B. subtilis flagellar filaments S17P | | Descriptor: | Flagellin | | Authors: | Wang, F, Burrage, A.M, Orlova, A, Kearns, D.B, Egelman, E.H. | | Deposit date: | 2017-07-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | A structural model of flagellar filament switching across multiple bacterial species.
Nat Commun, 8, 2017
|
|
5WJY
 
 | |
5WJZ
 
 | | Cryo-EM structure of B. subtilis flagellar filaments E115G | | Descriptor: | Flagellin | | Authors: | Wang, F, Burrage, A.M, Orlova, A, Kearns, D.B, Egelman, E.H. | | Deposit date: | 2017-07-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | A structural model of flagellar filament switching across multiple bacterial species.
Nat Commun, 8, 2017
|
|
5WK5
 
 | |
5WK6
 
 | |
5WLC
 
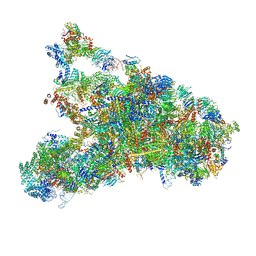 | | The complete structure of the small subunit processome | | Descriptor: | 18S pre-rRNA, 5' ETS, Bms1, ... | | Authors: | Barandun, J, Chaker-Margot, M, Hunziker, M, Klinge, S. | | Deposit date: | 2017-07-26 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The complete structure of the small-subunit processome.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5WP9
 
 | | Structural Basis of Mitochondrial Receptor Binding and Constriction by Dynamin-Related Protein 1 | | Descriptor: | Dynamin-1-like protein, MAGNESIUM ION, Mitochondrial dynamics protein MID49, ... | | Authors: | Kalia, R, Wang, R.Y.R, Yusuf, A, Thomas, P.V, Agard, D.A, Shaw, J.M, Frost, A. | | Deposit date: | 2017-08-03 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | Structural basis of mitochondrial receptor binding and constriction by DRP1.
Nature, 558, 2018
|
|
5WVI
 
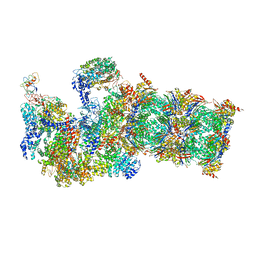 | | The resting state of yeast proteasome | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Ding, Z, Cong, Y. | | Deposit date: | 2016-12-25 | | Release date: | 2017-03-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | High-resolution cryo-EM structure of the proteasome in complex with ADP-AlFx
Cell Res., 27, 2017
|
|
5WVK
 
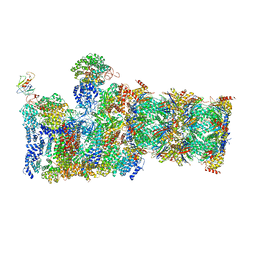 | | Yeast proteasome-ADP-AlFx | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Ding, Z, Cong, Y. | | Deposit date: | 2016-12-25 | | Release date: | 2017-03-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | High-resolution cryo-EM structure of the proteasome in complex with ADP-AlFx
Cell Res., 27, 2017
|
|
5WYJ
 
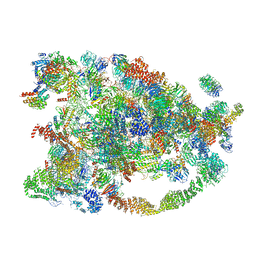 | | Cryo-EM structure of the 90S small subunit pre-ribosome (Dhr1-depleted, Enp1-TAP, state 1) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S ribosomal RNA, 40S ribosomal protein S1-A, ... | | Authors: | Ye, K, Zhu, X, Sun, Q. | | Deposit date: | 2017-01-13 | | Release date: | 2017-03-29 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Molecular architecture of the 90S small subunit pre-ribosome.
Elife, 6, 2017
|
|
5WYK
 
 | | Cryo-EM structure of the 90S small subunit pre-ribosome (Mtr4-depleted, Enp1-TAP) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S ribosomal RNA, 40S ribosomal protein S1-A, ... | | Authors: | Ye, K, Zhu, X, Sun, Q. | | Deposit date: | 2017-01-13 | | Release date: | 2017-03-29 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Molecular architecture of the 90S small subunit pre-ribosome.
Elife, 6, 2017
|
|
5X8P
 
 | | Structure of the 70S chloroplast ribosome from spinach | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Ahmed, T, Shi, J, Bhushan, S. | | Deposit date: | 2017-03-03 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Unique localization of the plastid-specific ribosomal proteins in the chloroplast ribosome small subunit provides mechanistic insights into the chloroplastic translation
Nucleic Acids Res., 45, 2017
|
|
5XJC
 
 | | Cryo-EM structure of the human spliceosome just prior to exon ligation at 3.6 angstrom | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, X, Yan, C, Hang, J, Finci, I.L, Lei, J, Shi, Y. | | Deposit date: | 2017-04-30 | | Release date: | 2017-07-05 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | An Atomic Structure of the Human Spliceosome
Cell, 169, 2017
|
|
5XTH
 
 | | Cryo-EM structure of human respiratory supercomplex I1III2IV1 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Acyl carrier protein, ... | | Authors: | Gu, J, Wu, M, Yang, M. | | Deposit date: | 2017-06-19 | | Release date: | 2017-09-13 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Architecture of Human Mitochondrial Respiratory Megacomplex I2III2IV2.
Cell, 170, 2017
|
|







