8S0K
 
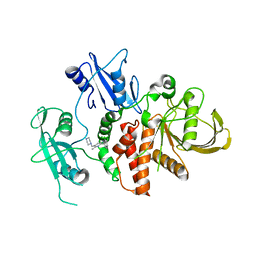 | | A fragment-based inhibitor of SHP2 | | Descriptor: | 3-[2,3-bis(chloranyl)phenyl]-5-methyl-6-(piperazin-1-ylmethyl)-1H-pyrrolo[3,2-b]pyridine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Cleasby, A, Price, A. | | Deposit date: | 2024-02-14 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Fragment-Based Discovery of Allosteric Inhibitors of SH2 Domain-Containing Protein Tyrosine Phosphatase-2 (SHP2).
J.Med.Chem., 67, 2024
|
|
8S0O
 
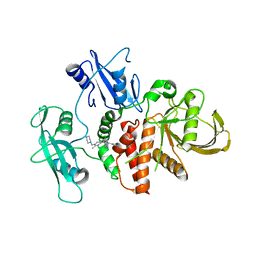 | | A fragment-based inhibitor of SHP2 | | Descriptor: | 3-[4-chloranyl-2-(1H-pyrazol-4-ylmethyl)indazol-5-yl]-5-methyl-6-(piperazin-1-ylmethyl)-1H-pyrrolo[3,2-b]pyridine, FORMIC ACID, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Cleasby, A, Price, A. | | Deposit date: | 2024-02-14 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.834 Å) | | Cite: | Fragment-Based Discovery of Allosteric Inhibitors of SH2 Domain-Containing Protein Tyrosine Phosphatase-2 (SHP2).
J.Med.Chem., 67, 2024
|
|
8S0P
 
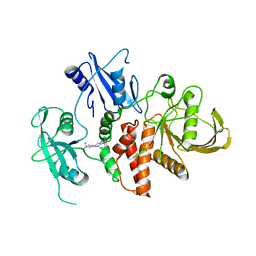 | | A fragment-based inhibitor of SHP2 | | Descriptor: | 1-[3-[2,3-bis(chloranyl)phenyl]-1H-pyrrolo[3,2-b]pyridin-6-yl]-4-methyl-piperidin-4-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Cleasby, A, Price, A. | | Deposit date: | 2024-02-14 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-Based Discovery of Allosteric Inhibitors of SH2 Domain-Containing Protein Tyrosine Phosphatase-2 (SHP2).
J.Med.Chem., 67, 2024
|
|
8S0Q
 
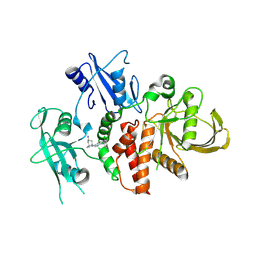 | | A fragment-based inhibitor of SHP2 | | Descriptor: | (1S,5R)-8-[3-[2,3-bis(chloranyl)phenyl]-1H-pyrrolo[3,2-b]pyridin-6-yl]-8-azabicyclo[3.2.1]octan-3-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Cleasby, A, Price, A. | | Deposit date: | 2024-02-14 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.872 Å) | | Cite: | Fragment-Based Discovery of Allosteric Inhibitors of SH2 Domain-Containing Protein Tyrosine Phosphatase-2 (SHP2).
J.Med.Chem., 67, 2024
|
|
8S0S
 
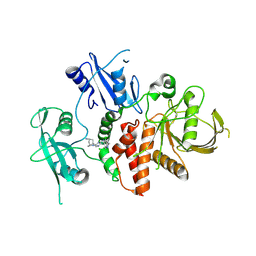 | | A fragment-based inhibitor of SHP2 | | Descriptor: | (1R,5S)-8-[7-(4-chloranyl-2-methyl-indazol-5-yl)-5H-pyrrolo[2,3-b]pyrazin-3-yl]-8-azabicyclo[3.2.1]octan-3-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Cleasby, A, Price, A. | | Deposit date: | 2024-02-14 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Fragment-Based Discovery of Allosteric Inhibitors of SH2 Domain-Containing Protein Tyrosine Phosphatase-2 (SHP2).
J.Med.Chem., 67, 2024
|
|
8S1K
 
 | | Crystal Structure of human FABP4 in complex with 2-[1-(methoxymethyl)cyclopentyl]-6-pentyl-4-phenyl-3-(1H-tetrazol-5-yl)-5,6,7,8-tetrahydroquinoline | | Descriptor: | (6~{S})-2-[1-(methoxymethyl)cyclopentyl]-6-pentyl-4-phenyl-3-(1~{H}-1,2,3,4-tetrazol-5-yl)-5,6,7,8-tetrahydroquinoline, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst-Sander, U, Rudolph, M.G. | | Deposit date: | 2024-02-15 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
8S1R
 
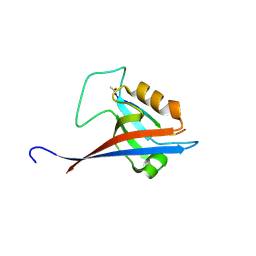 | |
8S2E
 
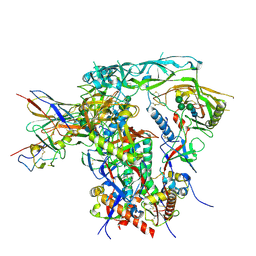 | | Fab4251-DS-SOSIP complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Nortier, P, Perez, L. | | Deposit date: | 2024-02-17 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | RAIN: a Machine Learning-based identification for HIV-1 bNAbs.
Res Sq, 2024
|
|
8S3B
 
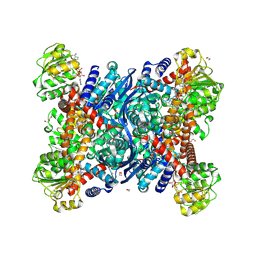 | | Crystal structure of Medicago truncatula glutamate dehydrogenase 2 in complex with 3-(1H-Tetrazol-5-yl)benzoic acid and NAD | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(1~{H}-1,2,3,4-tetrazol-5-yl)benzoic acid, ... | | Authors: | Grzechowiak, M, Ruszkowski, M. | | Deposit date: | 2024-02-19 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Legume-type glutamate dehydrogenase: Structure, activity, and inhibition studies.
Int.J.Biol.Macromol., 278, 2024
|
|
8S3R
 
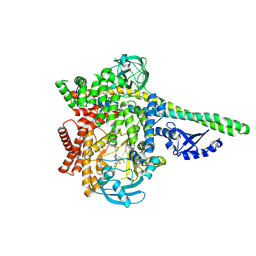 | | HUMAN PI3KDELTA IN COMPLEX WITH PYRIDAZINONE INHIBITOR 7 | | Descriptor: | 5-[(1~{S})-1-[4-azanyl-3-(5-oxidanylpyridin-3-yl)pyrazolo[3,4-d]pyrimidin-1-yl]ethyl]-4-cyclopentyl-2-(phenylmethyl)pyridazin-3-one, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Pala, D, Bruno, P, Capelli, A.M, Biagetti, M. | | Deposit date: | 2024-02-20 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery and Optimization of Pyridazinones as PI3K delta Selective Inhibitors for Administration by Inhalation.
J.Med.Chem., 67, 2024
|
|
8S3X
 
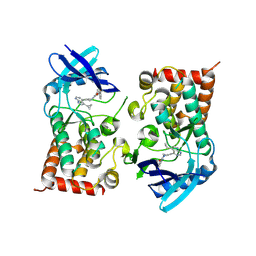 | | LIM Domain Kinase 2 (LIMK2) bound to compound 52 | | Descriptor: | 4-(5-cyclopropyl-7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl)-~{N}-[3-(3-methoxyphenyl)phenyl]-3,6-dihydro-2~{H}-pyridine-1-carboxamide, LIM domain kinase 2 | | Authors: | Mathea, S, Chatterjee, D, Preuss, F, Ple, K, Knapp, S. | | Deposit date: | 2024-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Tetrahydropyridine LIMK inhibitors: Structure activity studies and biological characterization.
Eur.J.Med.Chem., 271, 2024
|
|
8S51
 
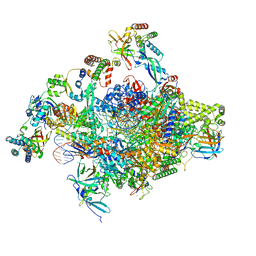 | | RNA polymerase II core initially transcribing complex with an ordered RNA of 8 nt | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8S54
 
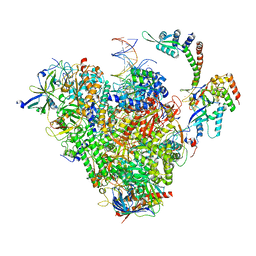 | | RNA polymerase II early elongation complex bound to TFIIE and TFIIF - state b (composite structure) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabber, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8S5B
 
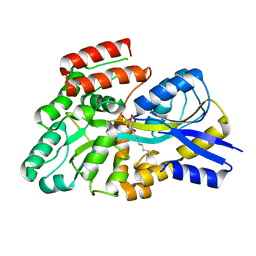 | | Crystal structure of the sulfoquinovosyl binding protein (SmoF) from A. tumefaciens sulfo-SMO pathway in complex with SQOctyl ligand | | Descriptor: | Sulfoquinovosyl glycerol-binding protein SmoF, [(2~{S},3~{S},4~{S},5~{R},6~{S})-6-octoxy-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Snow, A.J.D, Sharma, M, Davies, G.J. | | Deposit date: | 2024-02-23 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Capture-and-release of a sulfoquinovose-binding protein on sulfoquinovose-modified agarose.
Org.Biomol.Chem., 22, 2024
|
|
8S5Z
 
 | |
8S62
 
 | |
8S6H
 
 | |
8S7F
 
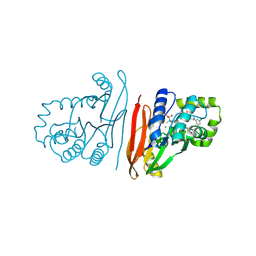 | | Crystal structure of Escherichia coli LpxH in complex with EBL-2805 | | Descriptor: | MANGANESE (II) ION, N-[4-[4-[3,5-bis(chloranyl)phenyl]piperazin-1-yl]sulfonylphenyl]-2-[methyl(methylsulfonyl)amino]benzamide, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Sooriyaarachchi, S, Mowbray, S.L, Bergfors, T, Jones, T.A. | | Deposit date: | 2024-03-01 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design, synthesis, and in vitro biological evaluation of meta-sulfonamidobenzamide-based antibacterial LpxH inhibitors.
Eur.J.Med.Chem., 278, 2024
|
|
8S7J
 
 | | Coxsackievirus A9 bound with compound 20 (CL300) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Plavec, Z, Butcher, S.J, Mitchell, C, Buckner, C. | | Deposit date: | 2024-03-01 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | SAR Analysis of Novel Coxsackie virus A9 Capsid Binders.
J.Med.Chem., 67, 2024
|
|
8S85
 
 | | Crystal structure of JAK1 JH1 domain in complex with an inhibitor | | Descriptor: | SODIUM ION, Tyrosine-protein kinase JAK1, ~{N}-(2-ethylphenyl)-~{N},1-dimethyl-imidazo[4,5-c]pyridin-6-amine | | Authors: | Flower, T.F, Lopez-Ramos, M, Geney, R, Jestel, A. | | Deposit date: | 2024-03-05 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design of a potent and selective dual JAK1/TYK2 inhibitor.
Bioorg.Med.Chem., 114, 2024
|
|
8S8C
 
 | | Structure of Kras in complex with inhibitor MK-1084 | | Descriptor: | (5aSa,17aRa)- 20-Chloro-2-[(2S,5R)-2,5-dimethyl-4-(prop-2-enoyl)piperazin-1-yl]-14,17-difluoro-6-(propan-2-yl)-11,12-dihydro-4H-1,18-(ethanediylidene)pyrido[4,3-e]pyrimido[1,6-g][1,4,7,9]benzodioxadiazacyclododecin-4-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Day, P.J, Cleasby, A. | | Deposit date: | 2024-03-06 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of MK-1084: An Orally Bioavailable and Low-Dose KRAS G12C Inhibitor.
J.Med.Chem., 67, 2024
|
|
8S9S
 
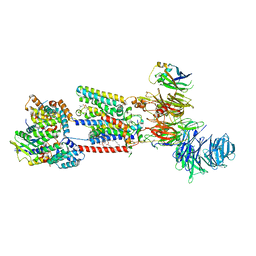 | | Structure of the human ER membrane protein complex (EMC) in GDN | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tomaleri, G.P, Nguyen, V.N, Voorhees, R.M. | | Deposit date: | 2023-03-30 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A selectivity filter in the ER membrane protein complex limits protein misinsertion at the ER.
J.Cell Biol., 222, 2023
|
|
8SAU
 
 | | CryoEM structure of DH270.4-CH848.10.17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848.10.17 gp120, CH848.10.17 gp41, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-02 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
8SAZ
 
 | | CryoEM structure of DH270.I5.6-CH848.10.17 | | Descriptor: | CH848.10.17.SOSIP gp41, DH270.I5.6 variable heavy chain, DH270.I5.6 variable light chain, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-02 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
8SB4
 
 | | CryoEM structure of DH270.1-CH848.10.17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848.10.17 gp120, CH848.10.17 gp41, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-02 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|






