9IIX
 
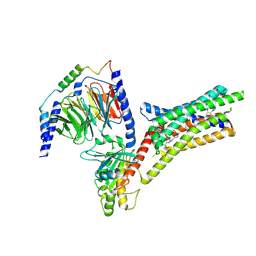 | | A Cryo-EM structure of Bitter taste receptor TAS2R14 with Ggust | | Descriptor: | 4-methyl-N-[(2M)-2-(1H-tetrazol-5-yl)phenyl]-6-(trifluoromethyl)pyrimidin-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yuan, Q, Duan, J, Tao, L, Xu, E.H. | | Deposit date: | 2024-06-21 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Bitter taste receptor TAS2R14 activation and G protein assembly by an intracellular agonist.
Cell Res., 2024
|
|
9IJ9
 
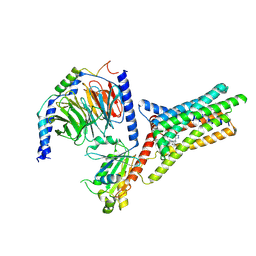 | | A Cryo-EM structure of Bitter taste receptor TAS2R14 with Gi complex | | Descriptor: | 4-methyl-N-[(2M)-2-(1H-tetrazol-5-yl)phenyl]-6-(trifluoromethyl)pyrimidin-2-amine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yuan, Q, Duan, J, Tao, L, Xu, E.H. | | Deposit date: | 2024-06-21 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Bitter taste receptor TAS2R14 activation and G protein assembly by an intracellular agonist.
Cell Res., 2024
|
|
9IJA
 
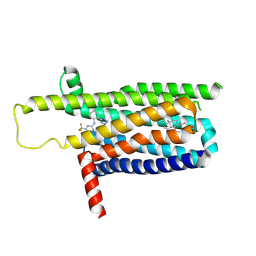 | | A local Cryo-EM structure of Bitter taste receptor TAS2R14 with Gi complex | | Descriptor: | 4-methyl-N-[(2M)-2-(1H-tetrazol-5-yl)phenyl]-6-(trifluoromethyl)pyrimidin-2-amine, CHOLESTEROL, Taste receptor type 2 member 14 | | Authors: | Yuan, Q, Duan, J, Tao, L, Xu, E.H. | | Deposit date: | 2024-06-21 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Bitter taste receptor TAS2R14 activation and G protein assembly by an intracellular agonist.
Cell Res., 2024
|
|
9IJC
 
 | |
9IK1
 
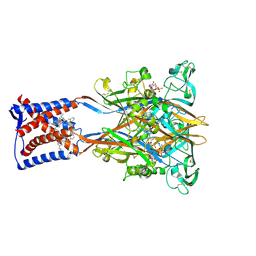 | | Cryo-EM structure of the human P2X3 receptor-compound 26a complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[2-cyclopropyl-7-[[(1~{R})-1-naphthalen-2-ylethyl]amino]-[1,2,4]triazolo[1,5-a]pyrimidin-5-yl]piperazine-1-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kim, S, Kim, G.R, Kim, Y.O, Han, X, Nagel, J, Kim, J, Song, D.I, Muller, C.E, Yoon, M.H, Jin, M.S, Kim, Y.C. | | Deposit date: | 2024-06-26 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Discovery of Triazolopyrimidine Derivatives as Selective P2X3 Receptor Antagonists Binding to an Unprecedented Allosteric Site as Evidenced by Cryo-Electron Microscopy.
J.Med.Chem., 67, 2024
|
|
9IK2
 
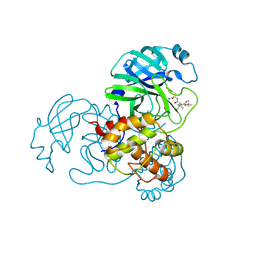 | | The co-crystal structure of SARS-CoV-2 Mpro in complex with compound H109 | | Descriptor: | 3C-like proteinase, tert-butyl N-[(2S)-1-[[(2S)-1-[[(2S)-1-azanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-1-oxidanylidene-3-phenyl-propan-2-yl]amino]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | Feng, Y, Zheng, W.Y, Han, P, Fu, L.F, Qi, J.X. | | Deposit date: | 2024-06-26 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided discovery of a small molecule inhibitor of SARS-CoV-2 main protease with potent in vitro and in vivo antiviral activities
To Be Published
|
|
9IMP
 
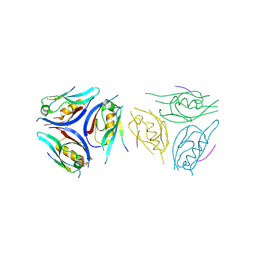 | | The complex of PDZ3 and PBM | | Descriptor: | INSC spindle orientation adaptor protein, Partitioning defective 3 homolog | | Authors: | Huang, S.J. | | Deposit date: | 2024-07-04 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | mInsc coordinates Par3 and NuMA condensates for assembly of the spindle orientation machinery in asymmetric cell division.
Int.J.Biol.Macromol., 279, 2024
|
|
9INR
 
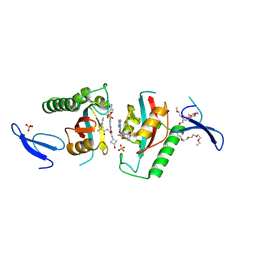 | | Crystal structure of PIN1 in complex with inhibitor C3 | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, SULFATE ION, ... | | Authors: | Zhang, L.Y. | | Deposit date: | 2024-07-08 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Re-Evaluating PIN1 as a Therapeutic Target in Oncology Using Neutral Inhibitors and PROTACs.
J.Med.Chem., 67, 2024
|
|
9INT
 
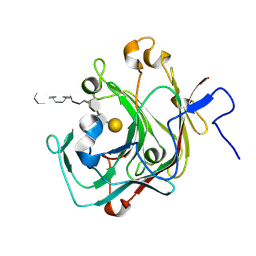 | | Crystal structure of the complex of the beta,kappa-carrageenase Cgbk16A from Wenyingzhuangia fucanilytica with an oligosaccharide of furcellaran | | Descriptor: | 3,6-anhydro-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-D-galactopyranose-(1-3)-beta-D-galactopyranose, GH16 domain-containing protein | | Authors: | Chang, Y, Chen, F. | | Deposit date: | 2024-07-08 | | Release date: | 2024-07-17 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural Insights into the Substrate Recognition and Catalytic Mechanism of a GH16 beta kappa-Carrageenase from Wenyingzhuangia fucanilytica.
J.Agric.Food Chem., 72, 2024
|
|
9INW
 
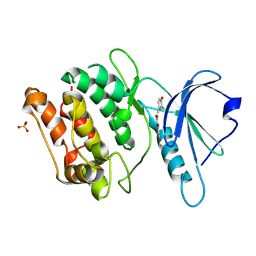 | | Crystal structure of DAPK1 in complex with compound 9 | | Descriptor: | (~{E})-1-[2,4-bis(oxidanyl)phenyl]-3-(3-chloranyl-4-oxidanyl-phenyl)prop-2-en-1-one, Death-associated protein kinase 1, SULFATE ION | | Authors: | Yokoyama, T. | | Deposit date: | 2024-07-08 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery and optimization of isoliquiritigenin as a death-associated protein kinase 1 inhibitor.
Eur.J.Med.Chem., 279, 2024
|
|
9INX
 
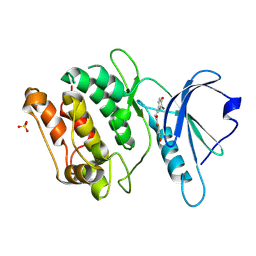 | | Crystal structure of DAPK1 in complex with compound 10 | | Descriptor: | (~{E})-1-[2,4-bis(oxidanyl)phenyl]-3-(3-bromanyl-4-oxidanyl-phenyl)prop-2-en-1-one, Death-associated protein kinase 1, SULFATE ION | | Authors: | Yokoyama, T. | | Deposit date: | 2024-07-08 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery and optimization of isoliquiritigenin as a death-associated protein kinase 1 inhibitor.
Eur.J.Med.Chem., 279, 2024
|
|
9INY
 
 | | Structure of bacteriophage T5 tail tube | | Descriptor: | Tail tube protein pb6 | | Authors: | Peng, Y.N, Liu, H.R. | | Deposit date: | 2024-07-08 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of Mature and Urea-Treated Empty Bacteriophage T5: Insights into Siphophage Infection and DNA Ejection.
Int J Mol Sci, 25, 2024
|
|
9IOZ
 
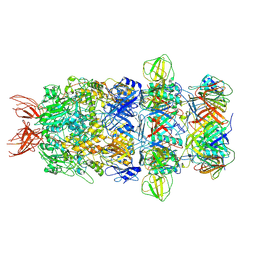 | | Structure of the bacteriophage T5 tail tip complex | | Descriptor: | Baseplate hub protein pb3, Baseplate tube protein p140, Distal tail protein pb9 | | Authors: | Peng, Y.N, Liu, H.R. | | Deposit date: | 2024-07-10 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of Mature and Urea-Treated Empty Bacteriophage T5: Insights into Siphophage Infection and DNA Ejection.
Int J Mol Sci, 25, 2024
|
|
9IUK
 
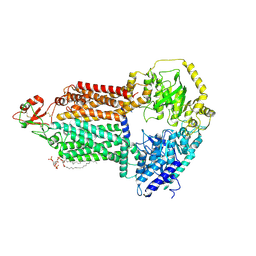 | | The structure of Candida albicans Cdr1 in apo state | | Descriptor: | Pip2(20:4/18:0), Pleiotropic ABC efflux transporter of multiple drugs CDR1 | | Authors: | Peng, Y, Sun, H, Yan, Z.F. | | Deposit date: | 2024-07-22 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structures of Candida albicans Cdr1 reveal azole-substrate recognition and inhibitor blocking mechanisms.
Nat Commun, 15, 2024
|
|
9IUL
 
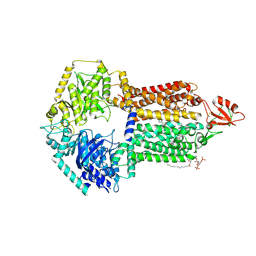 | | The structure of Candida albicans Cdr1 in fluconazole-bound state | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Pip2(20:4/18:0), Pleiotropic ABC efflux transporter of multiple drugs CDR1 | | Authors: | Peng, Y, Sun, H, Yan, Z.F. | | Deposit date: | 2024-07-23 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of Candida albicans Cdr1 reveal azole-substrate recognition and inhibitor blocking mechanisms.
Nat Commun, 15, 2024
|
|
9IUM
 
 | |
9J0U
 
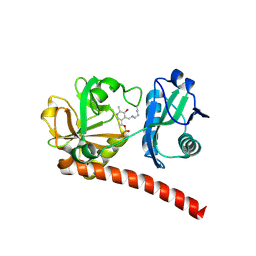 | | Crystal structure of monomeric PLP-dependent transaminase from Desulfobacula toluolica in F 41 3 2 space group | | Descriptor: | Dat: predicted D-alanine aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Bakunova, A.K, Nikolaeva, A.Y, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-08-03 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | High/low resolution monomeric PLP-dependent transaminase from Desulfobacula toluolica
To Be Published
|
|
9J4G
 
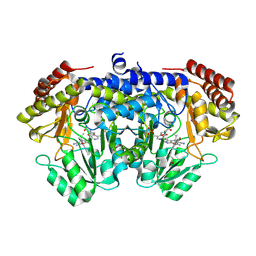 | | Crystal structure of SHMT from E. faecium with (+)-SHIN-2 | | Descriptor: | (+)-SHIN-2, Serine hydroxymethyltransferase, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-SERINE | | Authors: | Hayashi, H, Murayama, K. | | Deposit date: | 2024-08-09 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SHIN-2 exerts potent activity against VanA-type vancomycin-resistant Enterococcus faecium in vitro by stabilizing the active site loop of serine hydroxymethyltransferase.
Arch.Biochem.Biophys., 761, 2024
|
|
9J4L
 
 | | Crystal structure of GH9l Inulin fructotransferases (IFTase) | | Descriptor: | DFA-III-forming inulin fructotransferase | | Authors: | Chen, G, Wang, Z.X, Yang, Y.Q, Li, Y.G, Zhang, T, Ouyang, S.Y, Zhang, L, Chen, Y, Ruan, X.L, Miao, M. | | Deposit date: | 2024-08-09 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Elucidation of the mechanism underlying the sequential catalysis of inulin by fructotransferase.
Int.J.Biol.Macromol., 277, 2024
|
|






