9F5L
 
 | |
9F7A
 
 | |
9F7C
 
 | |
9F83
 
 | |
9FBG
 
 | |
8RJ7
 
 | |
8YKN
 
 | |
8YKP
 
 | |
9CRC
 
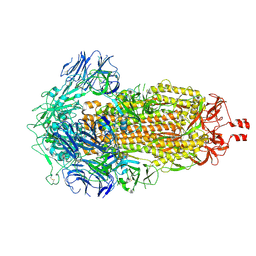 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: B.1 variant 3 closed RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ke, Z, Croll, T.I, Briggs, J.A.G. | | Deposit date: | 2024-07-22 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Virion morphology and on-virus spike protein structures of diverse SARS-CoV-2 variants.
Embo J., 43, 2024
|
|
9CRE
 
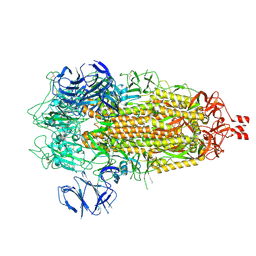 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: Alpha (B.1.1.7) variant 3 closed RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ke, Z, Croll, T.I, Briggs, J.A.G. | | Deposit date: | 2024-07-22 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Virion morphology and on-virus spike protein structures of diverse SARS-CoV-2 variants.
Embo J., 43, 2024
|
|
9CRG
 
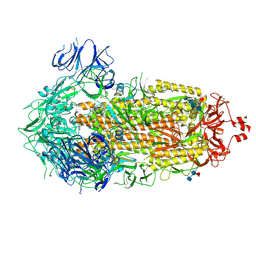 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: Gamma (P.1) variant 3 closed RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ke, Z, Kotecha, A, Briggs, J.A.G. | | Deposit date: | 2024-07-22 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Virion morphology and on-virus spike protein structures of diverse SARS-CoV-2 variants.
Embo J., 43, 2024
|
|
9CRH
 
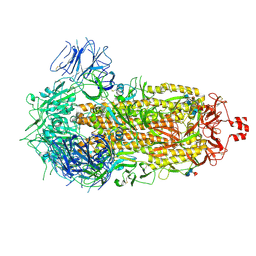 | |
9CRI
 
 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: Mu (B.1.621) variant 3 closed RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ke, Z, Briggs, J.A.G. | | Deposit date: | 2024-07-22 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Virion morphology and on-virus spike protein structures of diverse SARS-CoV-2 variants.
Embo J., 43, 2024
|
|
8R6E
 
 | | SARS-CoV-2 Nucleocapsid dimerization domain | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Fahoum, J, Wiener, R, Rouvinski, A, Isupov, M.N. | | Deposit date: | 2023-11-22 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | SARS-CoV-2 Nucleocapsid dimerization domain
To Be Published
|
|
8RNE
 
 | | HLA-E*01:03 in complex with SARS-CoV-2 Nsp13 peptide, VMPLSAPTL | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, E alpha chain variant, ... | | Authors: | Sun, R, Achour, A, Sala, B.M, Sandalova, T. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Emerging mutation in SARS-CoV-2 facilitates escape from NK cell recognition and associates with enhanced viral fitness.
Plos Pathog., 20, 2024
|
|
8RNF
 
 | | HLA-E*01:03 in complex with SARS-CoV-2 Omicron Nsp13 peptide, VIPLSAPTL | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, E alpha chain variant, ... | | Authors: | Sun, R, Achour, A, Sala, B.M, Sandalova, T. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.872 Å) | | Cite: | Emerging mutation in SARS-CoV-2 facilitates escape from NK cell recognition and associates with enhanced viral fitness.
Plos Pathog., 20, 2024
|
|
8USZ
 
 | | Cryo-EM Structure of Full-Length Spike Protein of Omicron XBB.1.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Huynh, K.W, Chang, J.S, Fennell, K.F, Che, Y, Wu, H. | | Deposit date: | 2023-10-30 | | Release date: | 2024-12-04 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Preclinical characterization of the Omicron XBB.1.5-adapted BNT162b2 COVID-19 vaccine.
Npj Vaccines, 9, 2024
|
|
8XKO
 
 | | CryoEM structure of compound HNC-1664 bound with RdRP-RNA complex of SARS-CoV-2 | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Li, M, An, L, Hong, Y, Li, S, Zhang, K. | | Deposit date: | 2023-12-23 | | Release date: | 2024-12-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | An adenosine analog shows high antiviral potency against coronavirus and arenavirus mainly through an unusual base pairing mode.
Nat Commun, 15, 2024
|
|
8YX5
 
 | | Crystal Structure of SARS CoV-2 Papain-like Protease PLpro-C111S in Complex with GZNL-P35 | | Descriptor: | 5-[(1~{R},5~{S})-3,6-diazabicyclo[3.1.1]heptan-3-yl]-2-methyl-~{N}-[1-(1-methyl-2-oxidanylidene-benzo[cd]indol-6-yl)cyclopropyl]benzamide, GLYCEROL, Papain-like protease nsp3, ... | | Authors: | Lu, Y, Shang, J. | | Deposit date: | 2024-04-02 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of orally bioavailable SARS-CoV-2 papain-like protease inhibitor as a potential treatment for COVID-19.
Nat Commun, 15, 2024
|
|
9B82
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody COVA2-15 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVA2-15 antibody heavy chain, COVA2-15 antibody light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2024-03-28 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Plant-produced SARS-CoV-2 antibody engineered towards enhanced potency and in vivo efficacy.
Plant Biotechnol J, 23, 2025
|
|
8RBY
 
 | |
8XEF
 
 | | Cocktail GC2050-GC2225 | | Descriptor: | GC2050 heavy chain, GC2050 light chain, GC2225 heavy chain, ... | | Authors: | Feng, L.L. | | Deposit date: | 2023-12-11 | | Release date: | 2024-12-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.41 Å) | | Cite: | XBB.1.5 RBD in complex with GC2050 and GC2225
To Be Published
|
|
9CJ6
 
 | |
8JAP
 
 | |
8V5V
 
 | | Structure of a SARS-CoV-2 spike S2 subunit in a pre-fusion, open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S2,Fibritin, ... | | Authors: | Olmedillas, E, Diaz, R, Hastie, K, Ollmann-Saphire, E. | | Deposit date: | 2023-12-01 | | Release date: | 2024-12-18 | | Last modified: | 2025-08-06 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structure of a SARS-CoV-2 spike S2 subunit in a pre-fusion, open conformation.
Cell Rep, 44, 2025
|
|








