8R6E
 
 | | SARS-CoV-2 Nucleocapsid dimerization domain | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Fahoum, J, Wiener, R, Rouvinski, A, Isupov, M.N. | | Deposit date: | 2023-11-22 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | SARS-CoV-2 Nucleocapsid dimerization domain
To Be Published
|
|
8RNE
 
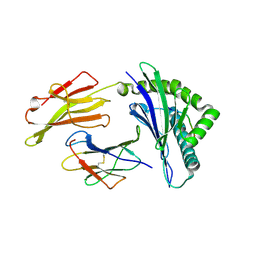 | | HLA-E*01:03 in complex with SARS-CoV-2 Nsp13 peptide, VMPLSAPTL | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, E alpha chain variant, ... | | Authors: | Sun, R, Achour, A, Sala, B.M, Sandalova, T. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Emerging mutation in SARS-CoV-2 facilitates escape from NK cell recognition and associates with enhanced viral fitness.
Plos Pathog., 20, 2024
|
|
8RNF
 
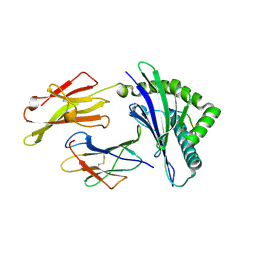 | | HLA-E*01:03 in complex with SARS-CoV-2 Omicron Nsp13 peptide, VIPLSAPTL | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, E alpha chain variant, ... | | Authors: | Sun, R, Achour, A, Sala, B.M, Sandalova, T. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.872 Å) | | Cite: | Emerging mutation in SARS-CoV-2 facilitates escape from NK cell recognition and associates with enhanced viral fitness.
Plos Pathog., 20, 2024
|
|
8USZ
 
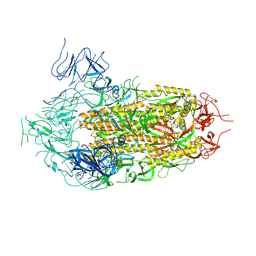 | | Cryo-EM Structure of Full-Length Spike Protein of Omicron XBB.1.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Huynh, K.W, Chang, J.S, Fennell, K.F, Che, Y, Wu, H. | | Deposit date: | 2023-10-30 | | Release date: | 2024-12-04 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Preclinical characterization of the Omicron XBB.1.5-adapted BNT162b2 COVID-19 vaccine.
Npj Vaccines, 9, 2024
|
|
8XKO
 
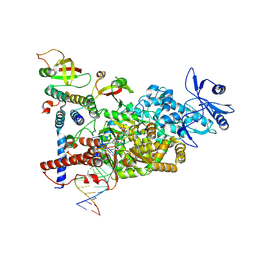 | | CryoEM structure of compound HNC-1664 bound with RdRP-RNA complex of SARS-CoV-2 | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Li, M, An, L, Hong, Y, Li, S, Zhang, K. | | Deposit date: | 2023-12-23 | | Release date: | 2024-12-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | An adenosine analog shows high antiviral potency against coronavirus and arenavirus mainly through an unusual base pairing mode.
Nat Commun, 15, 2024
|
|
8YX2
 
 | |
8YX3
 
 | |
8YX4
 
 | |
8YX5
 
 | | Crystal Structure of SARS CoV-2 Papain-like Protease PLpro-C111S in Complex with GZNL-P35 | | Descriptor: | 5-[(1~{R},5~{S})-3,6-diazabicyclo[3.1.1]heptan-3-yl]-2-methyl-~{N}-[1-(1-methyl-2-oxidanylidene-benzo[cd]indol-6-yl)cyclopropyl]benzamide, GLYCEROL, Papain-like protease nsp3, ... | | Authors: | Lu, Y, Shang, J. | | Deposit date: | 2024-04-02 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of orally bioavailable SARS-CoV-2 papain-like protease inhibitor as a potential treatment for COVID-19.
Nat Commun, 15, 2024
|
|
9B82
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody COVA2-15 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVA2-15 antibody heavy chain, COVA2-15 antibody light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2024-03-28 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Plant-produced SARS-CoV-2 antibody engineered towards enhanced potency and in vivo efficacy.
Plant Biotechnol J, 23, 2025
|
|
9GLV
 
 | | Crystal structure of SARS-CoV-2 Mpro with AB-343. | | Descriptor: | (1S,3S,4S)-N-[(2S)-1-azanylidene-3-[(3S)-5,5-dimethyl-2-oxidanylidene-pyrrolidin-3-yl]propan-2-yl]-2-[(2R)-3-cyclobutyl-2-[2,2,2-tris(fluoranyl)ethanoylamino]propanoyl]-5,5-bis(fluoranyl)-2-azabicyclo[2.2.2]octane-3-carboxamide, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Prasad, A, Blaesse, M, Maskos, K, Steinbacher, S, Konz Makino, D.L. | | Deposit date: | 2024-08-28 | | Release date: | 2024-12-04 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Biological characterization of AB-343, a novel and potent SARS-CoV-2 M pro inhibitor with pan-coronavirus activity.
Antiviral Res., 232, 2024
|
|
9GUD
 
 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and theophylline derivative LAS 54570922 | | Descriptor: | (3~{S})-3-azanyl-4-[(3~{R},4~{R},6~{S})-3-[1,3-dimethyl-2,6-bis(oxidanylidene)purin-7-yl]-4-methyl-4,6-bis(oxidanyl)azepan-1-yl]-4-oxidanylidene-butanoic acid, 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-09-19 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of SARS-CoV-2 methyltransferase nsp10-16 with Cap0-site binders
To Be Published
|
|
9GUY
 
 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and theophylline derivative LAS 54571098 | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-09-20 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of SARS-CoV-2 methyltransferase nsp10-16 with Cap0-site binders
To Be Published
|
|
9HAK
 
 | | Structure of compound 119 bound to SARS-CoV-2 main protease | | Descriptor: | (5~{R})-4-[(4-bromanyl-2-ethyl-phenyl)methyl]-~{N}-ethyl-1-thieno[2,3-c]pyridin-4-ylcarbonyl-1,4-diazepane-5-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Mac Sweeney, A, Hazemann, J. | | Deposit date: | 2024-11-04 | | Release date: | 2024-12-04 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Accelerating the Hit-To-Lead Optimization of a SARS-CoV-2 Mpro Inhibitor Series by Combining High-Throughput Medicinal Chemistry and Computational Simulations.
J.Med.Chem., 68, 2025
|
|
8RBY
 
 | |
8XEF
 
 | | Cocktail GC2050-GC2225 | | Descriptor: | GC2050 heavy chain, GC2050 light chain, GC2225 heavy chain, ... | | Authors: | Feng, L.L. | | Deposit date: | 2023-12-11 | | Release date: | 2024-12-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.41 Å) | | Cite: | XBB.1.5 RBD in complex with GC2050 and GC2225
To Be Published
|
|
9CJ6
 
 | |
9HJH
 
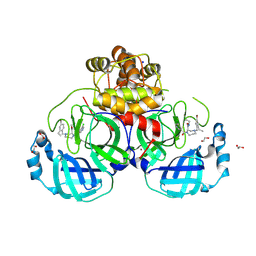 | | Structure of compound 1 bound to SARS-CoV-2 main protease | | Descriptor: | (2~{R})-4-[(4-bromanyl-2-ethyl-phenyl)methyl]-1-(5-chloranylpyridin-3-yl)carbonyl-~{N}-ethyl-1,4-diazepane-2-carboxamide, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Mac Sweeney, A, Hazemann, J. | | Deposit date: | 2024-11-29 | | Release date: | 2024-12-11 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Accelerating the Hit-To-Lead Optimization of a SARS-CoV-2 Mpro Inhibitor Series by Combining High-Throughput Medicinal Chemistry and Computational Simulations.
J.Med.Chem., 68, 2025
|
|
8JAP
 
 | |
8TYK
 
 | |
8V5V
 
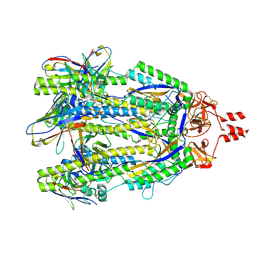 | | Structure of a SARS-CoV-2 spike S2 subunit in a pre-fusion, open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S2,Fibritin, ... | | Authors: | Olmedillas, E, Diaz, R, Hastie, K, Ollmann-Saphire, E. | | Deposit date: | 2023-12-01 | | Release date: | 2024-12-18 | | Last modified: | 2025-08-06 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structure of a SARS-CoV-2 spike S2 subunit in a pre-fusion, open conformation.
Cell Rep, 44, 2025
|
|
8XRQ
 
 | | SARS-CoV-2 BA.1 spike RBD in complex bound with VacBB-639 | | Descriptor: | Heavy chain of VacBB 639 Fab, Light chain of VacBB 639 Fab, Spike protein S1 | | Authors: | Liu, C.C, Ju, B, Zhang, Z. | | Deposit date: | 2024-01-08 | | Release date: | 2024-12-18 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | Rapid clonal expansion and somatic hypermutation contribute to the fate of SARS-CoV-2 broadly neutralizing antibodies.
J Immunol., 214, 2025
|
|
9BIH
 
 | | SARS-CoV-2 endoribonuclease Nsp15 bound to dsRNA with 1 nucleotide bulge | | Descriptor: | RNA (34-mer), RNA (35-mer), Uridylate-specific endoribonuclease nsp15 | | Authors: | Wright, Z.M, Butay, K.J, Krahn, J.M, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2024-04-23 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Spontaneous base flipping helps drive Nsp15's preferences in double stranded RNA substrates.
Nat Commun, 16, 2025
|
|
8UDF
 
 | | Crystal structure of SARS-CoV-2 3CL protease with inhibitor DEL_7 | | Descriptor: | 2-cyano-D-phenylalanyl-2,4-dichloro-N-[(2S)-1-(4-fluorophenyl)-4-(methylamino)-4-oxobutan-2-yl]-D-phenylalaninamide, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Forouhar, F, Liu, H, Zack, A, Iketani, S, Williams, A, Vaz, D.R, Habashi, D.L, Resnick, S.J, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2023-09-28 | | Release date: | 2024-12-25 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Development of small molecule non-covalent coronavirus 3CL protease inhibitors from DNA-encoded chemical library screening.
Nat Commun, 16, 2025
|
|
8UDJ
 
 | | Crystal structure of SARS-CoV-2 3CL protease with inhibitor DEL_2 | | Descriptor: | 2-cyano-D-phenylalanyl-2,4-dichloro-N-[(2S)-4-(methylamino)-4-oxo-1-phenylbutan-2-yl]-D-phenylalaninamide, 3C-like proteinase nsp5 | | Authors: | Forouhar, F, Liu, H, Zack, A, Iketani, S, Williams, A, Vaz, D.R, Habashi, D.L, Resnick, S.J, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2023-09-28 | | Release date: | 2024-12-25 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of small molecule non-covalent coronavirus 3CL protease inhibitors from DNA-encoded chemical library screening.
Nat Commun, 16, 2025
|
|








