8TFD
 
 | | Crystal structure of a stem-loop DNA aptamer complexed with SARS-CoV-2 nucleocapsid protein RNA-binding domain | | Descriptor: | 1,2-ETHANEDIOL, DNA aptamer, Nucleoprotein | | Authors: | Esler, M, Belica, C, Shi, K, Aihara, H. | | Deposit date: | 2023-07-10 | | Release date: | 2024-11-13 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A compact stem-loop DNA aptamer targets a uracil-binding pocket in the SARS-CoV-2 nucleocapsid RNA-binding domain.
Nucleic Acids Res., 52, 2024
|
|
8WUR
 
 | | Crystal structure of SARS-Cov-2 main protease D48N mutant in complex with shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase nsp5 | | Authors: | Zhao, Z.Y, Li, W.W, Zhang, J, Li, J. | | Deposit date: | 2023-10-21 | | Release date: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural Basis for the Inhibition of SARS-CoV-2 M pro D48N Mutant by Shikonin and PF-07321332.
Viruses, 16, 2023
|
|
8WYH
 
 | |
8WYJ
 
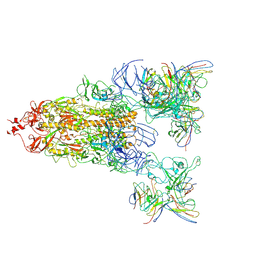 | | The global map of Omicron Subvariants Spike with two antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309-HC, S309-KC, ... | | Authors: | Yan, R.H, Wang, A.J, Yang, H.N. | | Deposit date: | 2023-10-31 | | Release date: | 2024-11-13 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the evolution and antibody evasion of SARS-CoV-2 BA.2.86 and JN.1 subvariants.
Nat Commun, 15, 2024
|
|
8XZ8
 
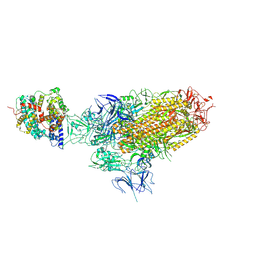 | | BA.2.86 Spike in complex with bovine ACE2 (bound 1 ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, CHLORIDE ION, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2024-01-21 | | Release date: | 2024-11-13 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | BA.2.86 Spike in complex with bovine ACE2
To Be Published
|
|
8XZ9
 
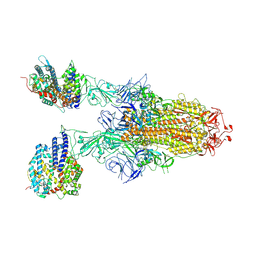 | | BA.2.86 Spike in complex with bovine ACE2 (bound 2 ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2024-01-21 | | Release date: | 2024-11-13 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | BA.2.86 Spike in complex with bovine ACE2
To Be Published
|
|
8XZA
 
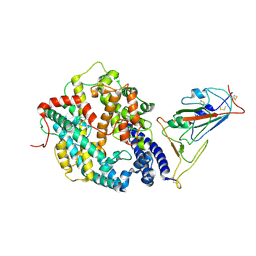 | |
8Y32
 
 | | BA.2.86-V483 RBD in complex with bACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, CHLORIDE ION, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2024-01-28 | | Release date: | 2024-11-13 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | BA.2.86-V483 RBD in complex with bACE2
To Be Published
|
|
9F2G
 
 | |
9F2H
 
 | | Crystal structure of SARS-CoV-2 N-protein C-terminal domain in complex with riluzole | | Descriptor: | 1,2-ETHANEDIOL, 6-(trifluoromethoxy)-1,3-benzothiazol-2-amine, Nucleoprotein, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B, Perez-Canadillas, J.M. | | Deposit date: | 2024-04-23 | | Release date: | 2024-11-13 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The ALS drug riluzole binds to the C-terminal domain of SARS-CoV-2 nucleocapsid protein and has antiviral activity.
Structure, 33, 2025
|
|
9F2I
 
 | | Crystal structure of SARS-CoV-2 N-protein C-terminal domain in complex with 2-amino-1,3-benzothiazol-6-ol | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-1,3-benzothiazol-6-ol, Nucleoprotein, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B, Perez-Canadillas, J.M. | | Deposit date: | 2024-04-23 | | Release date: | 2024-11-13 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The ALS drug riluzole binds to the C-terminal domain of SARS-CoV-2 nucleocapsid protein and has antiviral activity.
Structure, 33, 2025
|
|
8W1T
 
 | | SARS-CoV-2 Main protease bound to a non-covalent non-peptidic HTS hit | | Descriptor: | (5-bromopyridin-3-yl){4-[(R)-(4-chlorophenyl)(phenyl)methyl]piperazin-1-yl}methanone, (5-bromopyridin-3-yl){4-[(S)-(4-chlorophenyl)(phenyl)methyl]piperazin-1-yl}methanone, 1,2-ETHANEDIOL, ... | | Authors: | Bian, X, Tang, S, Sacchettini, J.C. | | Deposit date: | 2024-02-18 | | Release date: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An oral non-covalent non-peptidic inhibitor of SARS-CoV-2 Mpro ameliorates viral replication and pathogenesis in vivo.
Cell Rep, 43, 2024
|
|
8W1U
 
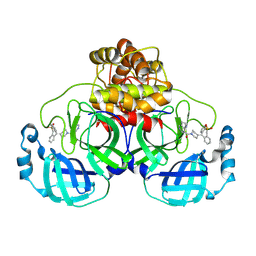 | | SARS-CoV-2 Main protease bound to non-covalent lead molecule NZ-804 | | Descriptor: | 11-[1-(1H-pyrrolo[3,2-c]pyridine-7-carbonyl)piperidin-4-ylidene]-6,11-dihydro-5H-5lambda~6~-dibenzo[b,e]thiepine-5,5-dione, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Bian, X, Tang, S, Sacchettini, J.C. | | Deposit date: | 2024-02-18 | | Release date: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | An oral non-covalent non-peptidic inhibitor of SARS-CoV-2 Mpro ameliorates viral replication and pathogenesis in vivo.
Cell Rep, 43, 2024
|
|
9C44
 
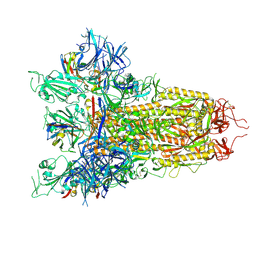 | |
9C45
 
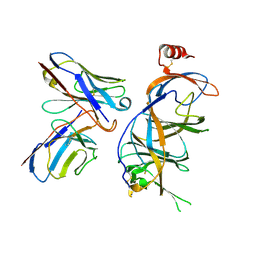 | |
9CO6
 
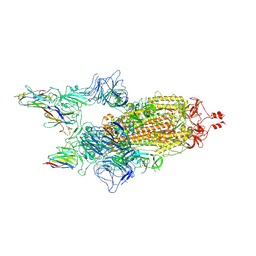 | | BA.5 spike/Nanosota-9 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-9, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2024-07-16 | | Release date: | 2024-11-20 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Discovery of Nanosota-9 as anti-Omicron nanobody therapeutic candidate.
Plos Pathog., 20, 2024
|
|
9CO7
 
 | | Local refinement of BA.5 spike/Nanosota-9 complex | | Descriptor: | Nanosota-9, Spike glycoprotein | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2024-07-16 | | Release date: | 2024-11-20 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Discovery of Nanosota-9 as anti-Omicron nanobody therapeutic candidate.
Plos Pathog., 20, 2024
|
|
9CO8
 
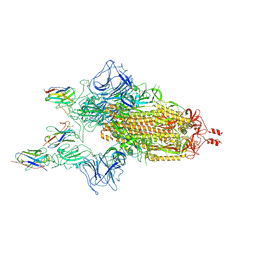 | | JN.1 spike/Nanosota-9 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-9, Spike glycoprotein | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2024-07-16 | | Release date: | 2024-11-20 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Discovery of Nanosota-9 as anti-Omicron nanobody therapeutic candidate.
Plos Pathog., 20, 2024
|
|
9CO9
 
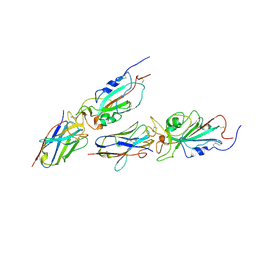 | | Local refinement of JN.1 spike/Nanosota-9 complex | | Descriptor: | Nanosota-9, Spike glycoprotein | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2024-07-16 | | Release date: | 2024-11-20 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Discovery of Nanosota-9 as anti-Omicron nanobody therapeutic candidate.
Plos Pathog., 20, 2024
|
|
9EVY
 
 | |
9EWH
 
 | |
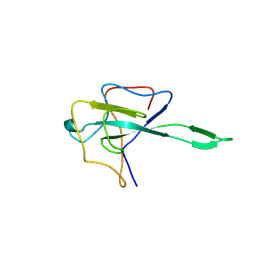
9EXB
 
 | |
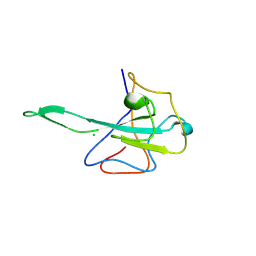
9EZB
 
 | |
9F5J
 
 | |
9F5L
 
 | |








