8YF2
 
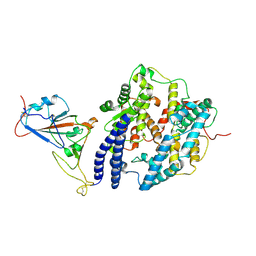 | | Cryo-EM structure of SARS-CoV-2 prototype RBD in complex with raccoon dog ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Li, L.J, Luo, C.L, Qi, J.X, Gao, G.F. | | Deposit date: | 2024-02-23 | | Release date: | 2024-10-23 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Receptor binding and structural basis of raccoon dog ACE2 binding to SARS-CoV-2 prototype and its variants.
Plos Pathog., 20, 2024
|
|
8YFT
 
 | | Cryo-EM structure of SARS-CoV-2 alpha variant spike protein in complex with raccoon dog ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Li, L.J, Luo, C.L, Qi, J.X, Gao, G.F. | | Deposit date: | 2024-02-25 | | Release date: | 2024-10-23 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Receptor binding and structural basis of raccoon dog ACE2 binding to SARS-CoV-2 prototype and its variants.
Plos Pathog., 20, 2024
|
|
9DW6
 
 | |
8RRN
 
 | |
9GDY
 
 | | SARS-CoV-2 Spike protein Beta Variant at 37C structural flexibility / heterogeneity analyses | | Descriptor: | Spike glycoprotein,Fibritin | | Authors: | Herreros, D, Mata, C.P, Noddings, C, Irene, D, Agard, D.A, Tsai, M.-D, Sorzano, C.O.S, Carazo, J.M. | | Deposit date: | 2024-08-06 | | Release date: | 2024-10-30 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Real-space heterogeneous reconstruction, refinement, and disentanglement of CryoEM conformational states with HetSIREN.
Nat Commun, 16, 2025
|
|
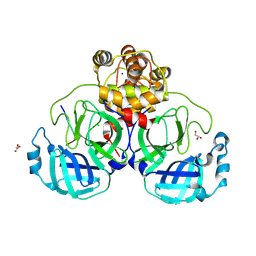
8U25
 
 | |
9CTU
 
 | | Cryo-EM structure of SARS-CoV-2 M (short conformation)bound to C1P | | Descriptor: | (2S,3R,4E)-2-(hexadecanoylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Membrane protein, POTASSIUM ION, ... | | Authors: | Dolan, K.A, Brohawn, S.G. | | Deposit date: | 2024-07-25 | | Release date: | 2024-11-06 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Direct lipid interactions control SARS-CoV-2 M protein conformational dynamics and virus assembly.
Biorxiv, 2024
|
|
9CTW
 
 | |
8R1Q
 
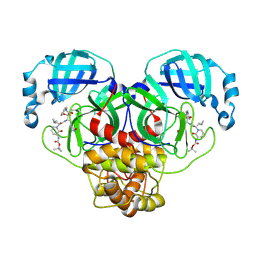 | | SARS-CoV-2 Mpro (Omicron, P132H+T169S) in complex with alpha-ketoamide 13b-K | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Sun, X, Ibrahim, M, Hilgenfeld, R. | | Deposit date: | 2023-11-02 | | Release date: | 2024-11-13 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Why is the Omicron main protease of SARS-CoV-2 less stable than its wild-type counterpart? A crystallographic, biophysical, and theoretical study
Hlife, 2, 2024
|
|
8TFD
 
 | | Crystal structure of a stem-loop DNA aptamer complexed with SARS-CoV-2 nucleocapsid protein RNA-binding domain | | Descriptor: | 1,2-ETHANEDIOL, DNA aptamer, Nucleoprotein | | Authors: | Esler, M, Belica, C, Shi, K, Aihara, H. | | Deposit date: | 2023-07-10 | | Release date: | 2024-11-13 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A compact stem-loop DNA aptamer targets a uracil-binding pocket in the SARS-CoV-2 nucleocapsid RNA-binding domain.
Nucleic Acids Res., 52, 2024
|
|
8WUR
 
 | | Crystal structure of SARS-Cov-2 main protease D48N mutant in complex with shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase nsp5 | | Authors: | Zhao, Z.Y, Li, W.W, Zhang, J, Li, J. | | Deposit date: | 2023-10-21 | | Release date: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural Basis for the Inhibition of SARS-CoV-2 M pro D48N Mutant by Shikonin and PF-07321332.
Viruses, 16, 2023
|
|
8WYH
 
 | |
8WYJ
 
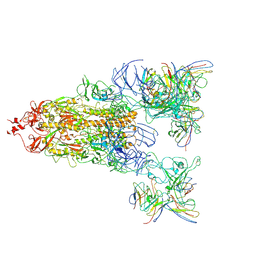 | |
8XZ8
 
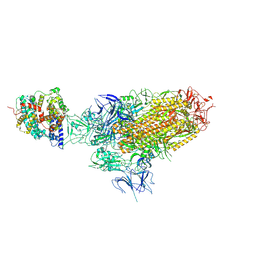 | | BA.2.86 Spike in complex with bovine ACE2 (bound 1 ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, CHLORIDE ION, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2024-01-21 | | Release date: | 2024-11-13 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | BA.2.86 Spike in complex with bovine ACE2
To Be Published
|
|
8XZ9
 
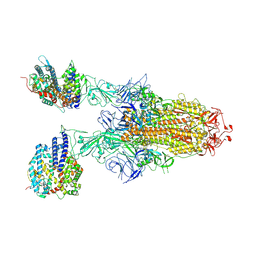 | | BA.2.86 Spike in complex with bovine ACE2 (bound 2 ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2024-01-21 | | Release date: | 2024-11-13 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | BA.2.86 Spike in complex with bovine ACE2
To Be Published
|
|
8XZA
 
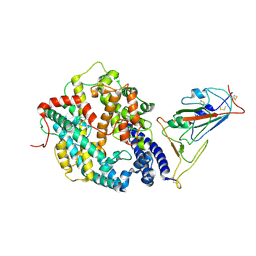 | |
8Y32
 
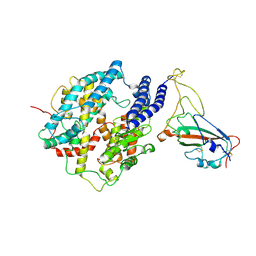 | | BA.2.86-V483 RBD in complex with bACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, CHLORIDE ION, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2024-01-28 | | Release date: | 2024-11-13 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | BA.2.86-V483 RBD in complex with bACE2
To Be Published
|
|
9F2H
 
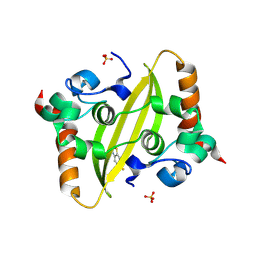 | | Crystal structure of SARS-CoV-2 N-protein C-terminal domain in complex with riluzole | | Descriptor: | 1,2-ETHANEDIOL, 6-(trifluoromethoxy)-1,3-benzothiazol-2-amine, Nucleoprotein, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B, Perez-Canadillas, J.M. | | Deposit date: | 2024-04-23 | | Release date: | 2024-11-13 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The ALS drug riluzole binds to the C-terminal domain of SARS-CoV-2 nucleocapsid protein and has antiviral activity.
Structure, 33, 2025
|
|
9C44
 
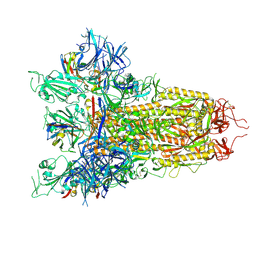 | |
9C45
 
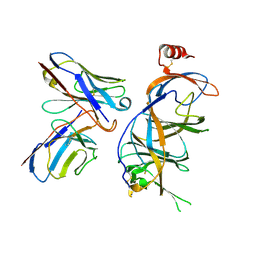 | |
9CO6
 
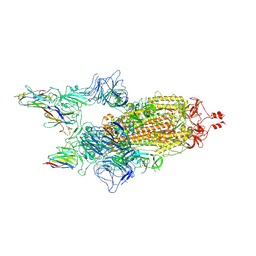 | | BA.5 spike/Nanosota-9 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-9, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2024-07-16 | | Release date: | 2024-11-20 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Discovery of Nanosota-9 as anti-Omicron nanobody therapeutic candidate.
Plos Pathog., 20, 2024
|
|
9CO7
 
 | | Local refinement of BA.5 spike/Nanosota-9 complex | | Descriptor: | Nanosota-9, Spike glycoprotein | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2024-07-16 | | Release date: | 2024-11-20 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Discovery of Nanosota-9 as anti-Omicron nanobody therapeutic candidate.
Plos Pathog., 20, 2024
|
|
9CO8
 
 | | JN.1 spike/Nanosota-9 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-9, Spike glycoprotein | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2024-07-16 | | Release date: | 2024-11-20 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Discovery of Nanosota-9 as anti-Omicron nanobody therapeutic candidate.
Plos Pathog., 20, 2024
|
|
9CO9
 
 | | Local refinement of JN.1 spike/Nanosota-9 complex | | Descriptor: | Nanosota-9, Spike glycoprotein | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2024-07-16 | | Release date: | 2024-11-20 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Discovery of Nanosota-9 as anti-Omicron nanobody therapeutic candidate.
Plos Pathog., 20, 2024
|
|
9EVY
 
 | |








