9BS8
 
 | |
9BSA
 
 | |
9BSE
 
 | |
9BSF
 
 | |
9BSG
 
 | |
9BSI
 
 | |
9BSO
 
 | |
9BSP
 
 | |
9BSQ
 
 | |
9BSR
 
 | |
9BST
 
 | |
9C2H
 
 | | SARS-CoV-2 Nucleocapsid Dimerization Domain bound to Fab-NP1E9 and Fab-NP3B4 | | Descriptor: | Antibody Fab NP1-E9 Heavy Chain (variable region), Antibody Fab NP1-E9 Light Chain (variable region), Antibody Fab NP3-B4 Heavy Chain (variable region), ... | | Authors: | Landeras-Bueno, S, Hariharan, C, Diaz Avalos, R, Ollmann Saphire, E. | | Deposit date: | 2024-05-31 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural stabilization of the intrinsically disordered SARS-CoV-2 N by binding to RNA sequences engineered from the viral genome
To Be Published
|
|
9F7P
 
 | |
9F7Q
 
 | |
9F7R
 
 | |
9F7S
 
 | |
9F7T
 
 | |
9F7U
 
 | |
9F7Y
 
 | |
9LVR
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, 6-(1,3-dihydroisoindol-2-yl)-3-(5-methylpyridin-3-yl)-1-[[3,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Unoh, Y, Hirai, K, Uehara, S, Kawashima, S, Nobori, H, Sato, J, Shibayama, H, Hori, A, Nakahara, K, Kurahashi, K, Takamatsu, M, Yamamoto, S, Zhang, O, Tanimura, M, Dodo, R, Maruyama, Y, Sawa, H, Watari, R, Miyano, T, Kato, T, Sato, T, Tachibana, Y. | | Deposit date: | 2025-02-12 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of The Clinical Candidate S-892216: A Second-Generation of SARS-CoV-2 3CL Protease Inhibitor for Treating COVID-19
To Be Published
|
|
9LVT
 
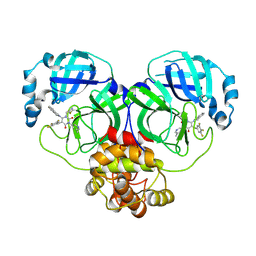 | | Crystal structure of SARS-CoV-2 3CL protease in complex with compound 4 | | Descriptor: | 1-(2-azanylideneethyl)-6-(1,3-dihydroisoindol-2-yl)-3-(5-methylpyridin-3-yl)-5-[[3,4,5-tris(fluoranyl)phenyl]methyl]pyrimidine-2,4-dione, 3C-like proteinase | | Authors: | Unoh, Y, Hirai, K, Uehara, S, Kawashima, S, Nobori, H, Sato, J, Shibayama, H, Hori, A, Nakahara, K, Kurahashi, K, Takamatsu, M, Yamamoto, S, Zhang, O, Tanimura, M, Dodo, R, Maruyama, Y, Sawa, H, Watari, R, Miyano, T, Kato, T, Sato, T, Tachibana, Y. | | Deposit date: | 2025-02-12 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of The Clinical Candidate S-892216: A Second-Generation of SARS-CoV-2 3CL Protease Inhibitor for Treating COVID-19
To Be Published
|
|
9LVV
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with compound 17 (S-892216) | | Descriptor: | 1-(2-azanylideneethyl)-6-[6,6-bis(fluoranyl)-2-azaspiro[3.3]heptan-2-yl]-5-(3-chloranyl-4-fluoranyl-phenyl)-3-(5-chloranylpyridin-3-yl)pyrimidine-2,4-dione, 3C-like proteinase | | Authors: | Unoh, Y, Hirai, K, Uehara, S, Kawashima, S, Nobori, H, Sato, J, Shibayama, H, Hori, A, Nakahara, K, Kurahashi, K, Takamatsu, M, Yamamoto, S, Zhang, O, Tanimura, M, Dodo, R, Maruyama, Y, Sawa, H, Watari, R, Miyano, T, Kato, T, Sato, T, Tachibana, Y. | | Deposit date: | 2025-02-12 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of The Clinical Candidate S-892216: A Second-Generation of SARS-CoV-2 3CL Protease Inhibitor for Treating COVID-19
To Be Published
|
|
7I13
 
 | | PanDDA analysis group deposition -- Main Protease (SARS-CoV-2) in complex with fragment B05 from the F2X-Entry Screen in orthorhombic space group | | Descriptor: | 2-acetylbenzoic acid, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Barthel, T, Benz, L.S, Wollenhaupt, J, Weiss, M.S. | | Deposit date: | 2025-02-03 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Orthorhombic SARS-CoV-2 main protease crystals provide higher success rate in fragment screening
To be published
|
|
7I14
 
 | | PanDDA analysis group deposition -- Main Protease (SARS-CoV-2) in complex with fragment B08 from the F2X-Entry Screen in orthorhombic space group | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ethyl 1,3-dihydro-2H-pyrrolo[3,4-c]pyridine-2-carboxylate | | Authors: | Barthel, T, Benz, L.S, Wollenhaupt, J, Weiss, M.S. | | Deposit date: | 2025-02-03 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Orthorhombic SARS-CoV-2 main protease crystals provide higher success rate in fragment screening
To be published
|
|
7I15
 
 | | PanDDA analysis group deposition -- Main Protease (SARS-CoV-2) in complex with fragment C02 from the F2X-Entry Screen in orthorhombic space group | | Descriptor: | 2,4,5-tris(fluoranyl)-3-methoxy-benzoic acid, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Barthel, T, Benz, L.S, Wollenhaupt, J, Weiss, M.S. | | Deposit date: | 2025-02-03 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Orthorhombic SARS-CoV-2 main protease crystals provide higher success rate in fragment screening
To be published
|
|








