7S5Q
 
 | |
7S5R
 
 | |
7TGE
 
 | |
7TGW
 
 | | Omicron spike at 3.0 A (open form) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Ye, G, Liu, B, Li, F. | | 登録日 | 2022-01-09 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Cryo-EM structure of a SARS-CoV-2 omicron spike protein ectodomain.
Nat Commun, 13, 2022
|
|
7TGX
 
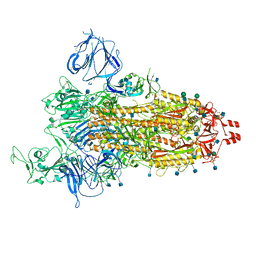 | | Prototypic SARS-CoV-2 G614 spike (open form) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose, ... | | 著者 | Ye, G, Liu, B, Li, F. | | 登録日 | 2022-01-09 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structure of a SARS-CoV-2 omicron spike protein ectodomain.
Nat Commun, 13, 2022
|
|
7TGY
 
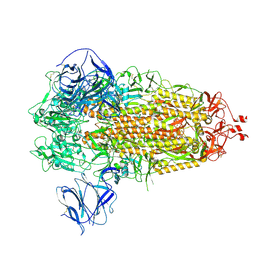 | | Prototypic SARS-CoV-2 G614 spike (closed form) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Ye, G, Liu, B, Li, F. | | 登録日 | 2022-01-09 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Cryo-EM structure of a SARS-CoV-2 omicron spike protein ectodomain.
Nat Commun, 13, 2022
|
|
7U28
 
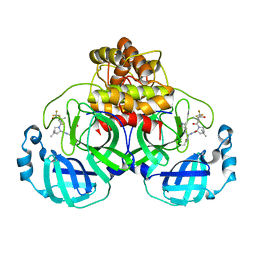 | | Structure of SARS-CoV-2 Mpro Lambda (G15S) in complex with Nirmatrelvir (PF-07321332) | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Greasley, S.E, Ferre, R.A, Plotnikova, O, Liu, W, Stewart, A.E. | | 登録日 | 2022-02-23 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.679 Å) | | 主引用文献 | Structural basis for the in vitro efficacy of nirmatrelvir against SARS-CoV-2 variants.
J.Biol.Chem., 298, 2022
|
|
7U29
 
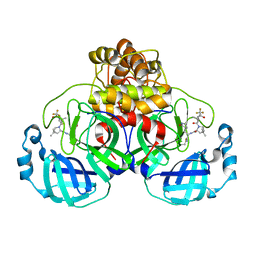 | | Structure of SARS-CoV-2 Mpro mutant (K90R) in complex with Nirmatrelvir (PF-07321332) | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | 登録日 | 2022-02-23 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.088 Å) | | 主引用文献 | Structural basis for the in vitro efficacy of nirmatrelvir against SARS-CoV-2 variants.
J.Biol.Chem., 298, 2022
|
|
7WP9
 
 | | SARS-CoV-2 Omicron Variant SPIKE trimer, all RBDs down | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | 登録日 | 2022-01-23 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (2.56 Å) | | 主引用文献 | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPA
 
 | | SARS-CoV-2 Omicron Variant SPIKE trimer complexed with ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | 登録日 | 2022-01-23 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (2.77 Å) | | 主引用文献 | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPB
 
 | | SARS-CoV-2 Omicron Variant RBD complexed with ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | 著者 | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | 登録日 | 2022-01-23 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (2.79 Å) | | 主引用文献 | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPC
 
 | | The second RBD of SARS-CoV-2 Omicron Variant in complexed with RBD-ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | 登録日 | 2022-01-23 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (2.57 Å) | | 主引用文献 | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
5SKW
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1272494722 | | 分子名称: | PHOSPHATE ION, Proofreading exoribonuclease nsp14, ZINC ION, ... | | 著者 | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | 登録日 | 2022-03-03 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.093 Å) | | 主引用文献 | PanDDA analysis group deposition
To Be Published
|
|
5SKX
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z126932614 | | 分子名称: | 2-[(methylsulfonyl)methyl]-1H-benzimidazole, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | 著者 | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | 登録日 | 2022-03-03 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.342 Å) | | 主引用文献 | PanDDA analysis group deposition
To Be Published
|
|
5SKY
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z466628048 | | 分子名称: | N-[(4-methyl-1,3-thiazol-2-yl)methyl]-1H-pyrazole-5-carboxamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | 著者 | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | 登録日 | 2022-03-03 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | PanDDA analysis group deposition
To Be Published
|
|
5SKZ
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z57258487 | | 分子名称: | N-{[4-(dimethylamino)phenyl]methyl}-4H-1,2,4-triazol-4-amine, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | 著者 | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | 登録日 | 2022-03-03 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | PanDDA analysis group deposition
To Be Published
|
|
5SL0
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z57260516 | | 分子名称: | 2-methoxy-~{N}-(2,4,6-trimethylphenyl)ethanamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | 著者 | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | 登録日 | 2022-03-03 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | PanDDA analysis group deposition
To Be Published
|
|
5SL1
 
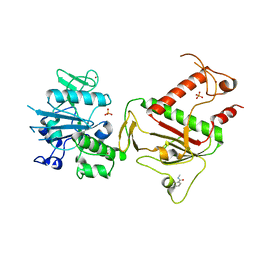 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1273312153 | | 分子名称: | N-methyl-1H-indole-7-carboxamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | 著者 | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | 登録日 | 2022-03-03 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.383 Å) | | 主引用文献 | PanDDA analysis group deposition
To Be Published
|
|
5SL2
 
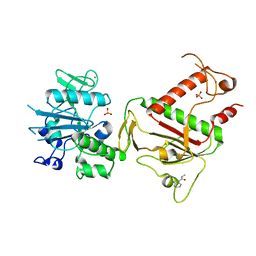 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z100643660 | | 分子名称: | N,1-dimethyl-1H-indole-3-carboxamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | 著者 | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | 登録日 | 2022-03-03 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.739 Å) | | 主引用文献 | PanDDA analysis group deposition
To Be Published
|
|
5SL3
 
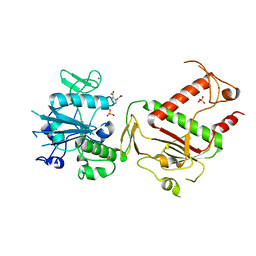 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z223688272 | | 分子名称: | 2-[acetyl(methyl)amino]benzoic acid, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | 著者 | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | 登録日 | 2022-03-03 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | PanDDA analysis group deposition
To Be Published
|
|
5SL4
 
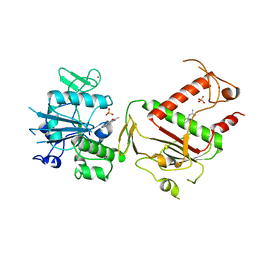 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z383202616 | | 分子名称: | N-(1H-indazol-6-yl)acetamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | 著者 | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | 登録日 | 2022-03-03 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.936 Å) | | 主引用文献 | PanDDA analysis group deposition
To Be Published
|
|
5SL5
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z32014663 | | 分子名称: | N,N,2,3-tetramethylbenzamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | 著者 | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | 登録日 | 2022-03-03 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | PanDDA analysis group deposition
To Be Published
|
|
5SL6
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z256709556 | | 分子名称: | 3-methylthiophene-2-carboxylic acid, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | 著者 | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | 登録日 | 2022-03-03 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.289 Å) | | 主引用文献 | PanDDA analysis group deposition
To Be Published
|
|
5SL7
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1186029914 | | 分子名称: | (3R)-1-(2-fluorophenyl)-3-(methylamino)pyrrolidin-2-one, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | 著者 | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | 登録日 | 2022-03-03 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.841 Å) | | 主引用文献 | PanDDA analysis group deposition
To Be Published
|
|
5SL8
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z2856434762 | | 分子名称: | N,N-dimethylpyridin-4-amine, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | 著者 | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | 登録日 | 2022-03-03 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | PanDDA analysis group deposition
To Be Published
|
|








