7GRU
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-17 | | 分子名称: | 3-(4-chlorophenyl)-1-methyl-1H-pyrazol-5-amine, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRV
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-18 | | 分子名称: | (2S)-2-(2-fluorophenyl)-1,3-thiazolidin-4-one, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRW
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-19 | | 分子名称: | (2S)-N-(3,5-dichlorophenyl)-2-hydroxypropanamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRX
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-20 | | 分子名称: | 1-(2,4-difluorophenyl)pyrrolidine-2,5-dione, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRY
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-21 | | 分子名称: | 1-(3,5-dichlorophenyl)pyrrolidine-2,5-dione, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRZ
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-22 | | 分子名称: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N,N-dimethyl-2-[(naphthalen-2-yl)oxy]acetamide, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GS0
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-23 | | 分子名称: | (pyridin-2-yl)(quinolin-2-yl)methanone, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GS1
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-24 | | 分子名称: | 2-cyano-~{N}-cyclohexyl-ethanamide, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GS2
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-25 | | 分子名称: | 3-(pyridin-3-yl)benzoic acid, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GS3
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-26 | | 分子名称: | (6-phenylpyridin-3-yl)methanamine, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GS4
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-27 | | 分子名称: | 3C-like proteinase nsp5, 7-(hydroxymethyl)-3-methyl-6~{H}-[1,3]thiazolo[3,2-a]pyrimidin-5-one, CHLORIDE ION, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GS5
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-28 | | 分子名称: | (3R)-5-fluoro-3-hydroxy-1,3-dihydro-2H-indol-2-one, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GS6
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-29 | | 分子名称: | (3S)-4,7-dichloro-3-hydroxy-1,3-dihydro-2H-indol-2-one, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RV4
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 2 | | 分子名称: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-2-phenyl-benzoic acid, ... | | 著者 | Kalnins, G. | | 登録日 | 2024-01-31 | | 公開日 | 2024-02-14 | | 最終更新日 | 2025-01-01 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural Basis for Inhibition of the SARS-CoV-2 nsp16 by Substrate-Based Dual Site Inhibitors.
Chemmedchem, 19, 2024
|
|
8RV5
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 1 | | 分子名称: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ... | | 著者 | Kalnins, G. | | 登録日 | 2024-01-31 | | 公開日 | 2024-02-14 | | 最終更新日 | 2025-01-01 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural Basis for Inhibition of the SARS-CoV-2 nsp16 by Substrate-Based Dual Site Inhibitors.
Chemmedchem, 19, 2024
|
|
8RV6
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 2 | | 分子名称: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-(4-hydroxyphenyl)benzoic acid, ... | | 著者 | Kalnins, G. | | 登録日 | 2024-01-31 | | 公開日 | 2024-02-14 | | 最終更新日 | 2025-01-01 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural Basis for Inhibition of the SARS-CoV-2 nsp16 by Substrate-Based Dual Site Inhibitors.
Chemmedchem, 19, 2024
|
|
8RV7
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 4 | | 分子名称: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-(3-oxidanylprop-1-ynyl)benzoic acid, ... | | 著者 | Kalnins, G. | | 登録日 | 2024-01-31 | | 公開日 | 2024-02-14 | | 最終更新日 | 2025-01-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Basis for Inhibition of the SARS-CoV-2 nsp16 by Substrate-Based Dual Site Inhibitors.
Chemmedchem, 19, 2024
|
|
8RV8
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 5 | | 分子名称: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]-2-chloranyl-benzoic acid, ... | | 著者 | Kalnins, G. | | 登録日 | 2024-01-31 | | 公開日 | 2024-02-14 | | 最終更新日 | 2025-01-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural Basis for Inhibition of the SARS-CoV-2 nsp16 by Substrate-Based Dual Site Inhibitors.
Chemmedchem, 19, 2024
|
|
8RV9
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 6 | | 分子名称: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-2-chloranyl-benzoic acid, ... | | 著者 | Kalnins, G. | | 登録日 | 2024-01-31 | | 公開日 | 2024-02-14 | | 最終更新日 | 2025-01-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Basis for Inhibition of the SARS-CoV-2 nsp16 by Substrate-Based Dual Site Inhibitors.
Chemmedchem, 19, 2024
|
|
8RVA
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 7 | | 分子名称: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]benzoic acid, ... | | 著者 | Kalnins, G. | | 登録日 | 2024-01-31 | | 公開日 | 2024-02-14 | | 最終更新日 | 2025-01-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Basis for Inhibition of the SARS-CoV-2 nsp16 by Substrate-Based Dual Site Inhibitors.
Chemmedchem, 19, 2024
|
|
8RVB
 
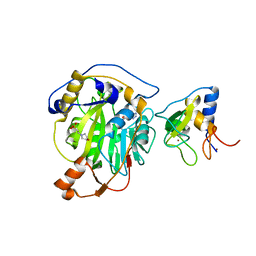 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 8 | | 分子名称: | (2~{R},3~{R},4~{S},5~{S})-2-(6-aminopurin-9-yl)-5-[2-(1~{H}-1,2,3-triazol-4-yl)ethylsulfanylmethyl]oxolane-3,4-diol, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | 著者 | Kalnins, G. | | 登録日 | 2024-01-31 | | 公開日 | 2024-02-14 | | 最終更新日 | 2025-01-01 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural Basis for Inhibition of the SARS-CoV-2 nsp16 by Substrate-Based Dual Site Inhibitors.
Chemmedchem, 19, 2024
|
|
8UIA
 
 | |
8UIF
 
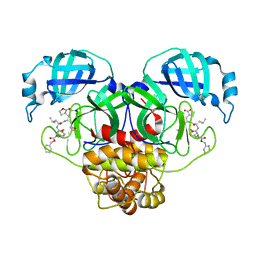 | |
8VKK
 
 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | 登録日 | 2024-01-09 | | 公開日 | 2024-02-14 | | 最終更新日 | 2025-05-28 | | 実験手法 | ELECTRON MICROSCOPY (2.81 Å) | | 主引用文献 | Altered receptor binding, antibody evasion and retention of T cell recognition by the SARS-CoV-2 XBB.1.5 spike protein.
Nat Commun, 15, 2024
|
|
8VKL
 
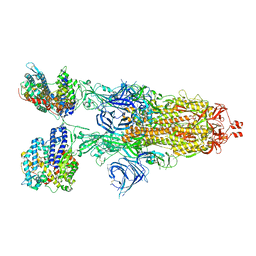 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (conformation 2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | 登録日 | 2024-01-09 | | 公開日 | 2024-02-14 | | 最終更新日 | 2025-02-26 | | 実験手法 | ELECTRON MICROSCOPY (2.91 Å) | | 主引用文献 | Altered receptor binding, antibody evasion and retention of T cell recognition by the SARS-CoV-2 XBB.1.5 spike protein.
Nat Commun, 15, 2024
|
|








