+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2ymn | ||||||
|---|---|---|---|---|---|---|---|
| Title | Organization of the Influenza Virus Replication Machinery | ||||||
 Components Components | NUCLEOPROTEIN | ||||||
 Keywords Keywords | VIRAL PROTEIN / RNP / POLYMERASE | ||||||
| Function / homology |  Function and homology information Function and homology informationhelical viral capsid / viral penetration into host nucleus / host cell / viral nucleocapsid / ribonucleoprotein complex / symbiont entry into host cell / host cell nucleus / structural molecule activity / RNA binding / identical protein binding Similarity search - Function | ||||||
| Biological species |   INFLUENZA A VIRUS INFLUENZA A VIRUS | ||||||
| Method | ELECTRON MICROSCOPY / helical reconstruction / cryo EM / Resolution: 20 Å | ||||||
 Authors Authors | Moeller, A. / Kirchdoerfer, R.N. / Potter, C.S. / Carragher, B. / Wilson, I.A. | ||||||
 Citation Citation |  Journal: Science / Year: 2012 Journal: Science / Year: 2012Title: Organization of the influenza virus replication machinery. Authors: Arne Moeller / Robert N Kirchdoerfer / Clinton S Potter / Bridget Carragher / Ian A Wilson /  Abstract: Influenza virus ribonucleoprotein complexes (RNPs) are central to the viral life cycle and in adaptation to new host species. RNPs are composed of the viral genome, viral polymerase, and many copies ...Influenza virus ribonucleoprotein complexes (RNPs) are central to the viral life cycle and in adaptation to new host species. RNPs are composed of the viral genome, viral polymerase, and many copies of the viral nucleoprotein. In vitro cell expression of all RNP protein components with four of the eight influenza virus gene segments enabled structural determination of native influenza virus RNPs by means of cryogenic electron microscopy (cryo-EM). The cryo-EM structure reveals the architecture and organization of the native RNP, defining the attributes of its largely helical structure and how polymerase interacts with nucleoprotein and the viral genome. Observations of branched-RNP structures in negative-stain electron microscopy and their putative identification as replication intermediates suggest a mechanism for viral replication by a second polymerase on the RNP template. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2ymn.cif.gz 2ymn.cif.gz | 465.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2ymn.ent.gz pdb2ymn.ent.gz | 382.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2ymn.json.gz 2ymn.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ym/2ymn https://data.pdbj.org/pub/pdb/validation_reports/ym/2ymn ftp://data.pdbj.org/pub/pdb/validation_reports/ym/2ymn ftp://data.pdbj.org/pub/pdb/validation_reports/ym/2ymn | HTTPS FTP |
|---|
-Related structure data
| Related structure data | 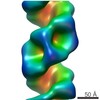 2209MC  2211MC  2212C  2213C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 56454.793 Da / Num. of mol.: 6 Source method: isolated from a genetically manipulated source Details: HELICAL ASSEMBLY OF NUCLEOPROTEINS WITHIN INFLUENZA RNP FILAMENT Source: (gene. exp.)  INFLUENZA A VIRUS (A/PUERTO RICO/8/1934(H1N1)) INFLUENZA A VIRUS (A/PUERTO RICO/8/1934(H1N1))Cell line (production host): HEK 293T17 / Production host:  HOMO SAPIENS (human) / References: UniProt: Q1I2B5, UniProt: Q1K9H2*PLUS HOMO SAPIENS (human) / References: UniProt: Q1I2B5, UniProt: Q1K9H2*PLUS |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: helical reconstruction |
- Sample preparation
Sample preparation
| Component | Name: INFLUENZA VIRUS RIBONUCLEOPROTEIN COMPLEX CENTRAL FILAMENT REGION Type: VIRUS |
|---|---|
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Specimen support | Details: HOLEY CARBON |
| Vitrification | Instrument: FEI VITROBOT MARK II / Cryogen name: ETHANE Details: VITRIFICATION 1 -- CRYOGEN- ETHANE, HUMIDITY- 85, INSTRUMENT- FEI VITROBOT MARK II, |
- Electron microscopy imaging
Electron microscopy imaging
| Microscopy | Model: FEI TECNAI 20 / Date: Aug 27, 2010 |
|---|---|
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 120 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 120 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 50000 X |
| Image recording | Electron dose: 15 e/Å2 / Film or detector model: GENERIC CCD |
| Radiation wavelength | Relative weight: 1 |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Details: WHOLEIMAGE | ||||||||||||||||||||||||||||||||
| 3D reconstruction | Method: IHRSR / Resolution: 20 Å / Num. of particles: 31573 / Nominal pixel size: 1.64 Å / Actual pixel size: 1.64 Å Details: SUBMISSION BASED ON EXPERIMENTAL DATA FROM EMDB EMD-2209.(DEPOSITION ID: 11133) Symmetry type: HELICAL | ||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: OTHER / Space: REAL / Details: REFINEMENT PROTOCOL--X-RAY | ||||||||||||||||||||||||||||||||
| Atomic model building | PDB-ID: 2IQH Accession code: 2IQH / Source name: PDB / Type: experimental model | ||||||||||||||||||||||||||||||||
| Refinement | Highest resolution: 20 Å | ||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Highest resolution: 20 Å
|
 Movie
Movie Controller
Controller









 PDBj
PDBj
