+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2213 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Organization of the Influenza Virus Replication Machinery | |||||||||
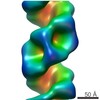
 Map data Map data | Reconstruction of Influenza Virus polymerase | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Influenza / RNP / Polymerase | |||||||||
| Biological species |  Influenza A virus (A/Puerto Rico/8/1934(H1N1)) Influenza A virus (A/Puerto Rico/8/1934(H1N1)) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 13.0 Å | |||||||||
 Authors Authors | Moeller A / Kirchdoerfer RN / Potter CS / Carragher B / Wilson IA | |||||||||
 Citation Citation |  Journal: Science / Year: 2012 Journal: Science / Year: 2012Title: Organization of the influenza virus replication machinery. Authors: Arne Moeller / Robert N Kirchdoerfer / Clinton S Potter / Bridget Carragher / Ian A Wilson /  Abstract: Influenza virus ribonucleoprotein complexes (RNPs) are central to the viral life cycle and in adaptation to new host species. RNPs are composed of the viral genome, viral polymerase, and many copies ...Influenza virus ribonucleoprotein complexes (RNPs) are central to the viral life cycle and in adaptation to new host species. RNPs are composed of the viral genome, viral polymerase, and many copies of the viral nucleoprotein. In vitro cell expression of all RNP protein components with four of the eight influenza virus gene segments enabled structural determination of native influenza virus RNPs by means of cryogenic electron microscopy (cryo-EM). The cryo-EM structure reveals the architecture and organization of the native RNP, defining the attributes of its largely helical structure and how polymerase interacts with nucleoprotein and the viral genome. Observations of branched-RNP structures in negative-stain electron microscopy and their putative identification as replication intermediates suggest a mechanism for viral replication by a second polymerase on the RNP template. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2213.map.gz emd_2213.map.gz | 7.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2213-v30.xml emd-2213-v30.xml emd-2213.xml emd-2213.xml | 7.8 KB 7.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_2213.tif emd_2213.tif | 185.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2213 http://ftp.pdbj.org/pub/emdb/structures/EMD-2213 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2213 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2213 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2213.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2213.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of Influenza Virus polymerase | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.18 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : influenza virus polymerase
| Entire | Name: influenza virus polymerase |
|---|---|
| Components |
|
-Supramolecule #1000: influenza virus polymerase
| Supramolecule | Name: influenza virus polymerase / type: sample / ID: 1000 / Number unique components: 3 |
|---|
-Macromolecule #1: influenza virus polymerase
| Macromolecule | Name: influenza virus polymerase / type: protein_or_peptide / ID: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/Puerto Rico/8/1934(H1N1)) Influenza A virus (A/Puerto Rico/8/1934(H1N1)) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 85 % / Instrument: HOMEMADE PLUNGER |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 20 |
|---|---|
| Date | Mar 11, 2011 |
| Image recording | Category: CCD / Film or detector model: GENERIC CCD / Number real images: 1878 / Average electron dose: 20 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal magnification: 100000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| CTF correction | Details: wholeimage |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 13.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: XMIPP, IMAGIC, SPIDER, APPION, CTFFIND3 / Number images used: 16958 |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















