+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDAK4 |
|---|---|
 Sample Sample | CRM1 RanGTP
|
| Function / homology |  Function and homology information Function and homology informationEstrogen-dependent nuclear events downstream of ESR-membrane signaling / Cyclin A/B1/B2 associated events during G2/M transition / HuR (ELAVL1) binds and stabilizes mRNA / Heme signaling / cellular response to triglyceride / cellular response to salt / Amplification of signal from unattached kinetochores via a MAD2 inhibitory signal / Deactivation of the beta-catenin transactivating complex / Transcriptional and post-translational regulation of MITF-M expression and activity / Mitotic Prometaphase ...Estrogen-dependent nuclear events downstream of ESR-membrane signaling / Cyclin A/B1/B2 associated events during G2/M transition / HuR (ELAVL1) binds and stabilizes mRNA / Heme signaling / cellular response to triglyceride / cellular response to salt / Amplification of signal from unattached kinetochores via a MAD2 inhibitory signal / Deactivation of the beta-catenin transactivating complex / Transcriptional and post-translational regulation of MITF-M expression and activity / Mitotic Prometaphase / EML4 and NUDC in mitotic spindle formation / Resolution of Sister Chromatid Cohesion / annulate lamellae / RHO GTPases Activate Formins / Separation of Sister Chromatids / regulation of proteasomal ubiquitin-dependent protein catabolic process / MAPK6/MAPK4 signaling / pre-miRNA export from nucleus / RNA nuclear export complex / snRNA import into nucleus / manchette / nuclear export signal receptor activity / regulation of centrosome duplication / cellular response to mineralocorticoid stimulus / Regulation of cholesterol biosynthesis by SREBP (SREBF) / importin-alpha family protein binding / regulation of protein export from nucleus / Rev-mediated nuclear export of HIV RNA / Nuclear import of Rev protein / protein localization to nucleolus / NEP/NS2 Interacts with the Cellular Export Machinery / tRNA processing in the nucleus / Postmitotic nuclear pore complex (NPC) reformation / GTP metabolic process / protein-containing complex localization / MicroRNA (miRNA) biogenesis / DNA metabolic process / regulation of protein catabolic process / dynein intermediate chain binding / mitotic sister chromatid segregation / viral process / ribosomal large subunit export from nucleus / spermatid development / protein localization to nucleus / nuclear pore / sperm flagellum / mRNA export from nucleus / Cajal body / ribosomal subunit export from nucleus / ribosomal small subunit export from nucleus / centriole / protein export from nucleus / mitotic spindle organization / male germ cell nucleus / hippocampus development / Transcriptional regulation by small RNAs / recycling endosome / positive regulation of protein import into nucleus / kinetochore / small GTPase binding / protein import into nucleus / GDP binding / melanosome / nuclear envelope / mitotic cell cycle / G protein activity / actin cytoskeleton organization / midbody / nuclear membrane / DNA-binding transcription factor binding / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / cadherin binding / response to xenobiotic stimulus / protein heterodimerization activity / ribonucleoprotein complex / protein domain specific binding / cell division / GTPase activity / chromatin binding / GTP binding / chromatin / protein-containing complex binding / nucleolus / magnesium ion binding / negative regulation of transcription by RNA polymerase II / protein-containing complex / RNA binding / extracellular exosome / nucleoplasm / membrane / nucleus / cytoplasm / cytosol Similarity search - Function |
| Biological species |   Homo sapiens (human) Homo sapiens (human) |
 Citation Citation |  Journal: Structure / Year: 2013 Journal: Structure / Year: 2013Title: Structural determinants and mechanism of mammalian CRM1 allostery. Authors: Nicole Dölker / Clement E Blanchet / Béla Voß / David Haselbach / Christian Kappel / Thomas Monecke / Dmitri I Svergun / Holger Stark / Ralf Ficner / Ulrich Zachariae / Helmut Grubmüller / Achim Dickmanns /  Abstract: Proteins carrying nuclear export signals cooperatively assemble with the export factor CRM1 and the effector protein RanGTP. In lower eukaryotes, this cooperativity is coupled to CRM1 conformational ...Proteins carrying nuclear export signals cooperatively assemble with the export factor CRM1 and the effector protein RanGTP. In lower eukaryotes, this cooperativity is coupled to CRM1 conformational changes; however, it is unknown if mammalian CRM1 maintains its compact conformation or shows similar structural flexibility. Here, combinations of small-angle X-ray solution scattering and electron microscopy experiments with molecular dynamics simulations reveal pronounced conformational flexibility in mammalian CRM1 and demonstrate that RanGTP binding induces association of its N- and C-terminal regions to form a toroid structure. The CRM1 toroid is stabilized mainly by local interactions between the terminal regions, rather than by global strain. The CRM1 acidic loop is key in transmitting the effect of this RanGTP-induced global conformational change to the NES-binding cleft by shifting its population to the open state, which displays enhanced cargo affinity. Cooperative CRM1 export complex assembly thus constitutes a highly dynamic process, encompassing an intricate interplay of global and local structural changes. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDAK4 SASDAK4 |
|---|
-Related structure data
- External links
External links
| Related items in Molecule of the Month |
|---|
-Models
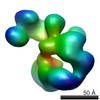
| Model #69 |  Type: dummy / Software: Dammif / Radius of dummy atoms: 1.90 A  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
|---|---|
| Model #72 |  Type: dummy / Software: Monsa / Radius of dummy atoms: 1.90 A / Chi-square value: 1.515361  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
| Model #73 |  Type: dummy / Software: Monsa / Radius of dummy atoms: 1.90 A / Chi-square value: 1.560001  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
- Sample
Sample
 Sample Sample | Name: CRM1 RanGTP / Sample MW: 147.51 kDa / Specimen concentration: 1.00-10.00 / Entity id: 62 / 63 |
|---|---|
| Buffer | Name: 50 mM Tris-HCL / pH: 7.5 / Composition: 150 mM NaCl, 1.0 mM DTT |
| Entity #62 | Type: protein / Description: Exportin-1 / Formula weight: 123.09 / Num. of mol.: 1 / Source: Mus musculus / References: UniProt: Q6P5F9 Sequence: MPAIMTMLAD HAARQLLDFS QKLDINLLDN VVNCLYHGEG AQQRMAQEVL THLKEHPDAW TRVDTILEFS QNMNTKYYGL QILENVIKTR WKILPRNQCE GIKKYVVGLI IKTSSDPTCV EKEKVYIGKL NMILVQILKQ EWPKHWPTFI SDIVGASRTS ESLCQNNMVI ...Sequence: MPAIMTMLAD HAARQLLDFS QKLDINLLDN VVNCLYHGEG AQQRMAQEVL THLKEHPDAW TRVDTILEFS QNMNTKYYGL QILENVIKTR WKILPRNQCE GIKKYVVGLI IKTSSDPTCV EKEKVYIGKL NMILVQILKQ EWPKHWPTFI SDIVGASRTS ESLCQNNMVI LKLLSEEVFD FSSGQITQVK AKHLKDSMCN EFSQIFQLCQ FVMENSQNAP LVHATLETLL RFLNWIPLGY IFETKLISTL IYKFLNVPMF RNVSLKCLTE IAGVSVSQYE EQFETLFTLT MMQLKQMLPL NTNIRLAYSN GKDDEQNFIQ NLSLFLCTFL KEHGQLLEKR LNLREALMEA LHYMLLVSEV EETEIFKICL EYWNHLAAEL YRESPFSTSA SPLLSGSQHF DIPPRRQLYL TVLSKVRLLM VSRMAKPEEV LVVENDQGEV VREFMKDTDS INLYKNMRET LVYLTHLDYV DTEIIMTKKL QNQVNGTEWS WKNLNTLCWA IGSISGAMHE EDEKRFLVTV IKDLLGLCEQ KRGKDNKAII ASNIMYIVGQ YPRFLRAHWK FLKTVVNKLF EFMHETHDGV QDMACDTFIK IAQKCRRHFV QVQVGEVMPF IDEILNNINT IICDLQPQQV HTFYEAVGYM IGAQTDQTVQ EHLIEKYMLL PNQVWDSIIQ QATKNVDILK DPETVKQLGS ILKTNVRACK AVGHPFVIQL GRIYLDMLNV YKCLSENISA AIQANGEMVT KQPLIRSMRT VKRETLKLIS GWVSRSNDPQ MVAENFVPPL LDAVLIDYQR NVPAAREPEV LSTMAIIVNK LGGHITAEIP QIFDAVFECT LNMINKDFEE YPEHRTNFFL LLQAVNSHCF PAFLAIPPAQ FKLVLDSIIW AFKHTMRNVA DTGLQILFTL LQNVAQEEAA AQSFYQTYFC DILQHIFSVV TDTSHTAGLT MHASILAYMF NLVEEGKIST PLNPGNPVNN QMFIQDYVAN LLKSAFPHLQ DAQVKLFVTG LFSLNQDIPA FKEHLRDFLV QIKEFAGEDT SDLFLEERET ALRQAQEEKH KLQMSVPGIL NPHEIPEEMC D |
| Entity #63 | Type: protein / Description: GTP-binding nuclear protein Ran / Formula weight: 24.42 / Num. of mol.: 1 / Source: Homo sapiens / References: UniProt: P62826 Sequence: MAAQGEPQVQ FKLVLVGDGG TGKTTFVKRH LTGEFEKKYV ATLGVEVHPL VFHTNRGPIK FNVWDTAGQE KFGGLRDGYY IQAQCAIIMF DVTSRVTYKN VPNWHRDLVR VCENIPIVLC GNKVDIKDRK VKAKSIVFHR KKNLQYYDIS AKSNYNFEKP FLWLARKLIG ...Sequence: MAAQGEPQVQ FKLVLVGDGG TGKTTFVKRH LTGEFEKKYV ATLGVEVHPL VFHTNRGPIK FNVWDTAGQE KFGGLRDGYY IQAQCAIIMF DVTSRVTYKN VPNWHRDLVR VCENIPIVLC GNKVDIKDRK VKAKSIVFHR KKNLQYYDIS AKSNYNFEKP FLWLARKLIG DPNLEFVAMP ALAPPEVVMD PALAAQYEHD LEVAQTTALP DEDDDL |
-Experimental information
| Beam | Instrument name:  DORIS III X33 DORIS III X33  / City: Hamburg / 国: Germany / City: Hamburg / 国: Germany  / Type of source: X-ray synchrotron / Type of source: X-ray synchrotron | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 2M | ||||||||||||||||||||||||
| Scan |
| ||||||||||||||||||||||||
| Result | Type of curve: single_conc /
|
 Movie
Movie Controller
Controller