+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDAK8 |
|---|---|
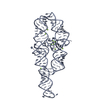
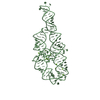
 Sample Sample | Dark state solution structure of Aureochrome1a- A´α-LOV-Jα
|
| Biological species |  |
 Citation Citation |  Journal: Structure / Year: 2016 Journal: Structure / Year: 2016Title: Structure of a Native-like Aureochrome 1a LOV Domain Dimer from Phaeodactylum tricornutum. Authors: Ankan Banerjee / Elena Herman / Tilman Kottke / Lars-Oliver Essen /  Abstract: Light-oxygen-voltage (LOV) domains absorb blue light for mediating various biological responses in all three domains of life. Aureochromes from stramenopile algae represent a subfamily of ...Light-oxygen-voltage (LOV) domains absorb blue light for mediating various biological responses in all three domains of life. Aureochromes from stramenopile algae represent a subfamily of photoreceptors that differs by its inversed topology with a C-terminal LOV sensor and an N-terminal effector (basic region leucine zipper, bZIP) domain. We crystallized the LOV domain including its flanking helices, A'α and Jα, of aureochrome 1a from Phaeodactylum tricornutum in the dark state and solved the structure at 2.8 Å resolution. Both flanking helices contribute to the interface of the native-like dimer. Small-angle X-ray scattering shows light-induced conformational changes limited to the dimeric envelope as well as increased flexibility in the lit state for the flanking helices. These rearrangements are considered to be crucial for the formation of the light-activated dimer. Finally, the LOV domain of the class 2 aureochrome PtAUREO2 was shown to lack a chromophore because of steric hindrance caused by M301. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Models
| Model #324 |  Type: dummy / Software: DAMMIF / Radius of dummy atoms: 3.25 A Comment: The model is an averaged and volume corrected representation (DAMFILT) Chi-square value: 0.595 / P-value: 0.000373  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
|---|
- Sample
Sample
 Sample Sample | Name: Dark state solution structure of Aureochrome1a- A´α-LOV-Jα Specimen concentration: 0.75-15.00 |
|---|---|
| Buffer | Name: Tris / Concentration: 10.00 mM / pH: 8 / Composition: 300 mM NaCl |
| Entity #191 | Name: Aureochrome1a-A / Type: protein / Description: Aureochrome1a-A´α-LOV-Jα / Formula weight: 17.973 / Num. of mol.: 2 / Source: Phaeodactylum tricornutum Sequence: MGSSHHHHHH SSGLVPRGSH MDFSFIKALQ TAQQNFVVTD PSLPDNPIVY ASQGFLNLTG YSLDQILGRN CRFLQGPETD PKAVERIRKA IEQGNDMSVC LLNYRVDGTT FWNQFFIAAL RDAGGNVTNF VGVQCKVSDQ YAATVTKQQE EEEEAAANDD ED |
-Experimental information
| Beam | Instrument name: ESRF BM29 / City: Grenoble / 国: France  / Type of source: X-ray synchrotron / Wavelength: 0.93 Å / Dist. spec. to detc.: 2.43 mm / Type of source: X-ray synchrotron / Wavelength: 0.93 Å / Dist. spec. to detc.: 2.43 mm | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 1M | |||||||||||||||||||||||||||
| Scan |
| |||||||||||||||||||||||||||
| Distance distribution function P(R) |
| |||||||||||||||||||||||||||
| Result | Comments: Solution scattering of N-terminally His6 tagged A´α-LOV-Jα was measured in the dark state (under safe red light). Two concentrations of the sample were measured at 40 and 833 micromolar ...Comments: Solution scattering of N-terminally His6 tagged A´α-LOV-Jα was measured in the dark state (under safe red light). Two concentrations of the sample were measured at 40 and 833 micromolar and then merged. Radiation damage was observed at very low extent. The data were normalized and the buffer only scattering subtracted. The analysis was performed using ATSAS. The displayed model is an averaged and volume corrected dummy atom representation (DAMFILT). For further sequence information refer to the Joint Genome Institute (JGI) entry JGI-49116.
|
 Movie
Movie Controller
Controller


 SASDAK8
SASDAK8