| Software | | Name | Version | Classification |
|---|
| HKL-2000 | | data scaling| PHENIX | 1.12_2829refinement| PDB_EXTRACT | 3.27 | data extraction| HKL-3000 | | data reduction| PHASER | | phasing | | | | | |
|
|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENT
Starting model: 5FFB
Resolution: 2.494→34.621 Å / SU ML: 0.38 / Cross valid method: THROUGHOUT / σ(F): 1.37 / Phase error: 29.12 / Stereochemistry target values: ML
| Rfactor | Num. reflection | % reflection |
|---|
| Rfree | 0.2484 | 388 | 4.81 % |
|---|
| Rwork | 0.203 | 7683 | - |
|---|
| obs | 0.2052 | 8071 | 85.41 % |
|---|
|
|---|
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL |
|---|
| Displacement parameters | Biso max: 111.37 Å2 / Biso mean: 52.2696 Å2 / Biso min: 30.33 Å2 |
|---|
| Refinement step | Cycle: final / Resolution: 2.494→34.621 Å
| Protein | Nucleic acid | Ligand | Solvent | Total |
|---|
| Num. atoms | 1375 | 0 | 5 | 38 | 1418 |
|---|
| Biso mean | - | - | 57.77 | 48.15 | - |
|---|
| Num. residues | - | - | - | - | 177 |
|---|
|
|---|
| Refine LS restraints | | Refine-ID | Type | Dev ideal | Number |
|---|
| X-RAY DIFFRACTION | f_bond_d| 0.004 | 1396 | | X-RAY DIFFRACTION | f_angle_d| 0.681 | 1855 | | X-RAY DIFFRACTION | f_chiral_restr| 0.038 | 187 | | X-RAY DIFFRACTION | f_plane_restr| 0.004 | 243 | | X-RAY DIFFRACTION | f_dihedral_angle_d| 15.923 | 883 | | | | | |
|
|---|
| Refine LS restraints NCS | | Ens-ID | Dom-ID | Auth asym-ID | Number | Refine-ID | Rms | Type |
|---|
| 1 | 1 | A| 796 | X-RAY DIFFRACTION | 10.137 | TORSIONAL| 1 | 2 | B| 796 | X-RAY DIFFRACTION | 10.137 | TORSIONAL | | | |
|
|---|
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Rfactor Rfree error: 0 | Resolution (Å) | Rfactor Rfree | Num. reflection Rfree | Rfactor Rwork | Num. reflection Rwork | % reflection obs (%) |
|---|
| 2.494-2.8544 | 0.3573 | 89 | 0.2806 | 1655 | 56 | | 2.8544-3.5956 | 0.2512 | 116 | 0.2312 | 3025 | 100 | | 3.5956-34.621 | 0.2261 | 183 | 0.1721 | 3003 | 99 |
|
|---|
| Refinement TLS params. | Method: refined / Origin x: 1.435 Å / Origin y: 23.7456 Å / Origin z: -3.4004 Å
| 11 | 12 | 13 | 21 | 22 | 23 | 31 | 32 | 33 |
|---|
| T | 0.7594 Å2 | 0.0717 Å2 | 0.0696 Å2 | - | 0.1508 Å2 | 0.0199 Å2 | - | - | 0.3372 Å2 |
|---|
| L | 1.159 °2 | -0.0705 °2 | 0.8054 °2 | - | 1.372 °2 | 0.1773 °2 | - | - | 1.7114 °2 |
|---|
| S | 0.119 Å ° | -0.1297 Å ° | -0.0268 Å ° | 0.2015 Å ° | 0.0349 Å ° | 0.0574 Å ° | 0.3675 Å ° | 0.0621 Å ° | -0.1094 Å ° |
|---|
|
|---|
| Refinement TLS group | | ID | Refine-ID | Refine TLS-ID | Selection details | Auth asym-ID | Auth seq-ID |
|---|
| 1 | X-RAY DIFFRACTION | 1 | allA| 32 - 116 | | 2 | X-RAY DIFFRACTION | 1 | allB| 25 - 116 | | 3 | X-RAY DIFFRACTION | 1 | allC| 1 - 2 | | 4 | X-RAY DIFFRACTION | 1 | allC| 3 - 4 | | 5 | X-RAY DIFFRACTION | 1 | allC| 5 | | 6 | X-RAY DIFFRACTION | 1 | allS| 1 - 39 | | | | | | | | | | | | |
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
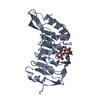
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.494 Å
MOLECULAR REPLACEMENT / Resolution: 2.494 Å  Authors
Authors China, 1items
China, 1items  Citation
Citation Journal: J.Inorg.Biochem. / Year: 2022
Journal: J.Inorg.Biochem. / Year: 2022 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 7vw1.cif.gz
7vw1.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb7vw1.ent.gz
pdb7vw1.ent.gz PDB format
PDB format 7vw1.json.gz
7vw1.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/vw/7vw1
https://data.pdbj.org/pub/pdb/validation_reports/vw/7vw1 ftp://data.pdbj.org/pub/pdb/validation_reports/vw/7vw1
ftp://data.pdbj.org/pub/pdb/validation_reports/vw/7vw1


 Links
Links Assembly
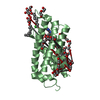
Assembly
 Components
Components

 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  SSRF
SSRF  / Beamline: BL19U1 / Wavelength: 0.9792 Å
/ Beamline: BL19U1 / Wavelength: 0.9792 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller










 PDBj
PDBj





