[English] 日本語
 Yorodumi
Yorodumi- PDB-7prc: PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS (DG-... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7prc | ||||||
|---|---|---|---|---|---|---|---|
| Title | PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS (DG-420315 (TRIAZINE) COMPLEX) | ||||||
 Components Components | (PHOTOSYNTHETIC REACTION ...) x 4 | ||||||
 Keywords Keywords | PHOTOSYNTHETIC REACTION CENTER / SECONDARY QUINONE (QB) / TRIAZINE INHIBITOR | ||||||
| Function / homology |  Function and homology information Function and homology informationplasma membrane-derived chromatophore membrane / plasma membrane light-harvesting complex / bacteriochlorophyll binding / : / photosynthetic electron transport in photosystem II / photosynthesis, light reaction / photosynthesis / electron transfer activity / iron ion binding / heme binding / metal ion binding Similarity search - Function | ||||||
| Biological species |  Blastochloris viridis (bacteria) Blastochloris viridis (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / DIFFERENCE FOURIER / SA OMIT MAPS / Resolution: 2.65 Å SYNCHROTRON / DIFFERENCE FOURIER / SA OMIT MAPS / Resolution: 2.65 Å | ||||||
 Authors Authors | Lancaster, C.R.D. / Michel, H. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1999 Journal: J.Mol.Biol. / Year: 1999Title: Refined crystal structures of reaction centres from Rhodopseudomonas viridis in complexes with the herbicide atrazine and two chiral atrazine derivatives also lead to a new model of the bound carotenoid. Authors: Lancaster, C.R. / Michel, H. #1:  Journal: Biochim.Biophys.Acta / Year: 1998 Journal: Biochim.Biophys.Acta / Year: 1998Title: Ubiquinone Reduction and Protonation in the Reaction Centre of Rhodopseudomonas Viridis: X-Ray Structures and Their Functional Implications Authors: Lancaster, C.R.D. #2:  Journal: Structure / Year: 1997 Journal: Structure / Year: 1997Title: The Coupling of Light-Induced Electron Transfer and Proton Uptake as Derived from Crystal Structures of Reaction Centres from Rhodopseudomonas Viridis Modified at the Binding Site of the Secondary Quinone, Qb Authors: Lancaster, C.R. / Michel, H. #3:  Journal: J.Mol.Biol. / Year: 1995 Journal: J.Mol.Biol. / Year: 1995Title: Crystallographic Refinement at 2.3 A Resolution and Refined Model of the Photosynthetic Reaction Centre from Rhodopseudomonas Viridis Authors: Deisenhofer, J. / Epp, O. / Sinning, I. / Michel, H. #4:  Journal: Science / Year: 1989 Journal: Science / Year: 1989Title: The Photosynthetic Reaction Center from the Purple Bacterium Rhodopseudomonas Viridis Authors: Deisenhofer, J. / Michel, H. #5:  Journal: Nature / Year: 1985 Journal: Nature / Year: 1985Title: Structure of the Protein Subunits in the Photosynthetic Reaction Centre of Rhodopseudomonas Viridis at 3 Angstroms Resolution Authors: Deisenhofer, J. / Epp, O. / Miki, K. / Huber, R. / Michel, H. #6:  Journal: J.Mol.Biol. / Year: 1984 Journal: J.Mol.Biol. / Year: 1984Title: X-Ray Structure Analysis of a Membrane Protein Complex. Electron Density Map at 3 A Resolution and a Model of the Chromophores of the Photosynthetic Reaction Center from Rhodopseudomonas Viridis Authors: Deisenhofer, J. / Epp, O. / Miki, K. / Huber, R. / Michel, H. #7:  Journal: J.Mol.Biol. / Year: 1982 Journal: J.Mol.Biol. / Year: 1982Title: Three-Dimensional Crystals of a Membrane Protein Complex. The Photosynthetic Reaction Centre from Rhodopseudomonas Viridis Authors: Michel, H. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7prc.cif.gz 7prc.cif.gz | 284.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7prc.ent.gz pdb7prc.ent.gz | 223.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7prc.json.gz 7prc.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/pr/7prc https://data.pdbj.org/pub/pdb/validation_reports/pr/7prc ftp://data.pdbj.org/pub/pdb/validation_reports/pr/7prc ftp://data.pdbj.org/pub/pdb/validation_reports/pr/7prc | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5prcC  6prcSC S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
-PHOTOSYNTHETIC REACTION ... , 4 types, 4 molecules CLMH
| #1: Protein | Mass: 37450.801 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Blastochloris viridis (bacteria) / Cellular location: INTRACYTOPLASMIC MEMBRANE (ICM) / References: UniProt: P07173 Blastochloris viridis (bacteria) / Cellular location: INTRACYTOPLASMIC MEMBRANE (ICM) / References: UniProt: P07173 |
|---|---|
| #2: Protein | Mass: 30469.104 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Blastochloris viridis (bacteria) / Cellular location: INTRACYTOPLASMIC MEMBRANE (ICM) / References: UniProt: P06009 Blastochloris viridis (bacteria) / Cellular location: INTRACYTOPLASMIC MEMBRANE (ICM) / References: UniProt: P06009 |
| #3: Protein | Mass: 35932.188 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Blastochloris viridis (bacteria) / Cellular location: INTRACYTOPLASMIC MEMBRANE (ICM) / References: UniProt: P06010 Blastochloris viridis (bacteria) / Cellular location: INTRACYTOPLASMIC MEMBRANE (ICM) / References: UniProt: P06010 |
| #4: Protein | Mass: 28557.453 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Blastochloris viridis (bacteria) / Cellular location: INTRACYTOPLASMIC MEMBRANE (ICM) / References: UniProt: P06008 Blastochloris viridis (bacteria) / Cellular location: INTRACYTOPLASMIC MEMBRANE (ICM) / References: UniProt: P06008 |
-Non-polymers , 10 types, 339 molecules 
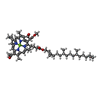

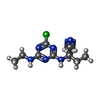















| #5: Chemical | ChemComp-HEM / #6: Chemical | ChemComp-BCB / #7: Chemical | #8: Chemical | ChemComp-CET / | #9: Chemical | ChemComp-LDA / #10: Chemical | ChemComp-FE2 / | #11: Chemical | ChemComp-SO4 / #12: Chemical | ChemComp-MQ7 / | #13: Chemical | ChemComp-NS5 / | #14: Water | ChemComp-HOH / | |
|---|
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 4 X-RAY DIFFRACTION / Number of used crystals: 4 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 4.9 Å3/Da / Density % sol: 70 % | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 6 / Details: pH 6.0 | ||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion | ||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 263 K | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: MPG/DESY, HAMBURG SYNCHROTRON / Site: MPG/DESY, HAMBURG  / Beamline: BW6 / Wavelength: 0.92 / Wavelength: 0.92, 1.0 / Beamline: BW6 / Wavelength: 0.92 / Wavelength: 0.92, 1.0 | |||||||||
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Aug 1, 1993 / Details: FOCUSSING MIRROR | |||||||||
| Radiation | Monochromator: GE(111), SI(111) / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||
| Radiation wavelength |
| |||||||||
| Reflection | Resolution: 2.6→30 Å / Num. obs: 76928 / % possible obs: 87 % / Redundancy: 4.6 % / Rmerge(I) obs: 0.074 / Rsym value: 0.074 / Net I/σ(I): 7.4 | |||||||||
| Reflection shell | Resolution: 2.6→2.67 Å / Redundancy: 1.7 % / Rmerge(I) obs: 0.39 / Mean I/σ(I) obs: 0.8 / Rsym value: 0.39 / % possible all: 71 | |||||||||
| Reflection | *PLUS Num. obs: 72314 / % possible obs: 88.3 % / Num. measured all: 352617 | |||||||||
| Reflection shell | *PLUS % possible obs: 77.8 % / Rmerge(I) obs: 0.279 / Mean I/σ(I) obs: 2.9 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure: DIFFERENCE FOURIER / SA OMIT MAPS Starting model: PDB ENTRY 6PRC Resolution: 2.65→10 Å / Rfactor Rfree error: 0.0027 / Cross valid method: A POSTERIORI / Details: N(OBS)/N(PAR) = 1.73
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 23.5 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.26 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.65→10 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.65→2.77 Å / Rfactor Rfree error: 0.015 / Total num. of bins used: 8
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Version: 3.1 / Classification: refinement X-PLOR / Version: 3.1 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor obs: 0.19 / Rfactor Rfree: 0.23 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS Biso mean: 23.2 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Rfactor obs: 0.334 |
 Movie
Movie Controller
Controller















 PDBj
PDBj



















