[English] 日本語
 Yorodumi
Yorodumi- PDB-7nay: Crystal structure of lactate dehydrogenase from Selenomonas rumin... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7nay | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of lactate dehydrogenase from Selenomonas ruminantium with NADH. | ||||||
 Components Components | L-lactate dehydrogenase | ||||||
 Keywords Keywords | OXIDOREDUCTASE / SELENOMONAS RUMINANTIUM / LACTATE DEHYDROGENASE / NADH | ||||||
| Function / homology |  Function and homology information Function and homology informationL-lactate dehydrogenase / L-lactate dehydrogenase (NAD+) activity / lactate metabolic process / cytoplasm Similarity search - Function | ||||||
| Biological species |  Selenomonas ruminantium (bacteria) Selenomonas ruminantium (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 1.84 Å MOLECULAR REPLACEMENT / Resolution: 1.84 Å | ||||||
 Authors Authors | Bertrand, Q. / Robin, A. / Girard, E. / Madern, D. | ||||||
| Funding support |  France, 1items France, 1items
| ||||||
 Citation Citation |  Journal: J.Struct.Biol. / Year: 2023 Journal: J.Struct.Biol. / Year: 2023Title: Biochemical, structural and dynamical characterizations of the lactate dehydrogenase from Selenomonas ruminantium provide information about an intermediate evolutionary step prior to complete ...Title: Biochemical, structural and dynamical characterizations of the lactate dehydrogenase from Selenomonas ruminantium provide information about an intermediate evolutionary step prior to complete allosteric regulation acquisition in the super family of lactate and malate dehydrogenases. Authors: Bertrand, Q. / Coquille, S. / Iorio, A. / Sterpone, F. / Madern, D. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7nay.cif.gz 7nay.cif.gz | 89 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7nay.ent.gz pdb7nay.ent.gz | 64.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7nay.json.gz 7nay.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  7nay_validation.pdf.gz 7nay_validation.pdf.gz | 751.7 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  7nay_full_validation.pdf.gz 7nay_full_validation.pdf.gz | 752.8 KB | Display | |
| Data in XML |  7nay_validation.xml.gz 7nay_validation.xml.gz | 17.2 KB | Display | |
| Data in CIF |  7nay_validation.cif.gz 7nay_validation.cif.gz | 25.8 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/na/7nay https://data.pdbj.org/pub/pdb/validation_reports/na/7nay ftp://data.pdbj.org/pub/pdb/validation_reports/na/7nay ftp://data.pdbj.org/pub/pdb/validation_reports/na/7nay | HTTPS FTP |
-Related structure data
| Related structure data |  8q3cC  3d0oS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||
| Unit cell |
| ||||||||||||
| Components on special symmetry positions |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 35925.844 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: Presence of 2 modified cysteines that are appearing during crystallisation process. Confirmed by Mass spectrometry on dissolved crystals. Source: (gene. exp.)  Selenomonas ruminantium (bacteria) / Gene: ldh, ldhL / Production host: Selenomonas ruminantium (bacteria) / Gene: ldh, ldhL / Production host:  |
|---|
-Non-polymers , 6 types, 300 molecules 

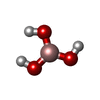








| #2: Chemical | ChemComp-NAD / | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| #3: Chemical | | #4: Chemical | #5: Chemical | ChemComp-EDO / #6: Chemical | #7: Water | ChemComp-HOH / | |
-Details
| Has ligand of interest | N |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.53 Å3/Da / Density % sol: 51.29 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 5 Details: 21 % PEG 1500 150 mM Malonate-Imidazole-Boric Acid pH 5 (MIB-PACT) 10 mM Sodium Bicarbonate pH 8 10 mM Tb-XO4 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source: SEALED TUBE / Type: Xenocs GeniX 3D Cu HF / Wavelength: 1.54179 Å |
| Detector | Type: MAR scanner 345 mm plate / Detector: IMAGE PLATE / Date: Dec 9, 2020 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.54179 Å / Relative weight: 1 |
| Reflection | Resolution: 1.84→38.61 Å / Num. obs: 31954 / % possible obs: 99.9 % / Redundancy: 5.6 % / CC1/2: 0.99 / Net I/σ(I): 12.17 |
| Reflection shell | Resolution: 1.84→1.95 Å / Redundancy: 5.3 % / Num. unique obs: 5035 / CC1/2: 0.83 / % possible all: 99.9 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3D0O Resolution: 1.84→38.61 Å / Cor.coef. Fo:Fc: 0.97 / Cor.coef. Fo:Fc free: 0.947 / SU B: 2.69 / SU ML: 0.079 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.117 / ESU R Free: 0.116 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS U VALUES : REFINED INDIVIDUALLY
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 75.68 Å2 / Biso mean: 18.2 Å2 / Biso min: 8.58 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 1.84→38.61 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.84→1.89 Å / Rfactor Rfree error: 0 / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller












 PDBj
PDBj








