[English] 日本語
 Yorodumi
Yorodumi- PDB-7l9g: Crystal structure of the second bromodomain (BD2) of human BRD2 b... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7l9g | ||||||
|---|---|---|---|---|---|---|---|
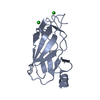
| Title | Crystal structure of the second bromodomain (BD2) of human BRD2 bound to BI2536 | ||||||
 Components Components | Bromodomain-containing protein 2 | ||||||
 Keywords Keywords | TRANSCRIPTION/TRANSCRIPTION INHIBITOR / BET / ERK5 / dual BRD-kinase inhibitor / GENE REGULATION / TRANSCRIPTION-TRANSCRIPTION INHIBITOR complex | ||||||
| Function / homology |  Function and homology information Function and homology informationhistone H4K12ac reader activity / histone H4K5ac reader activity / histone H3K14ac reader activity / acetylation-dependent protein binding / chromatin looping / RUNX3 regulates p14-ARF / positive regulation of T-helper 17 cell lineage commitment / protein localization to chromatin / neural tube closure / nucleosome assembly ...histone H4K12ac reader activity / histone H4K5ac reader activity / histone H3K14ac reader activity / acetylation-dependent protein binding / chromatin looping / RUNX3 regulates p14-ARF / positive regulation of T-helper 17 cell lineage commitment / protein localization to chromatin / neural tube closure / nucleosome assembly / histone binding / spermatogenesis / nuclear speck / chromatin remodeling / protein serine/threonine kinase activity / chromatin binding / regulation of transcription by RNA polymerase II / chromatin / nucleoplasm / nucleus / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.36 Å MOLECULAR REPLACEMENT / Resolution: 1.36 Å | ||||||
 Authors Authors | Karim, M.R. / Bikowitz, M.J. / Schonbrunn, E. | ||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2021 Journal: J.Med.Chem. / Year: 2021Title: Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors. Authors: Karim, R.M. / Bikowitz, M.J. / Chan, A. / Zhu, J.Y. / Grassie, D. / Becker, A. / Berndt, N. / Gunawan, S. / Lawrence, N.J. / Schonbrunn, E. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7l9g.cif.gz 7l9g.cif.gz | 89.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7l9g.ent.gz pdb7l9g.ent.gz | 66.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7l9g.json.gz 7l9g.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/l9/7l9g https://data.pdbj.org/pub/pdb/validation_reports/l9/7l9g ftp://data.pdbj.org/pub/pdb/validation_reports/l9/7l9g ftp://data.pdbj.org/pub/pdb/validation_reports/l9/7l9g | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5v67C  5vboC  5vbpC  5vbqC  5vbrC  7bjyC  7k6gC  7k6hC  7ko0C  7l6dC  7l72C  7l73C  7l9jC  7l9kC  7l9lC  7lahC  7laiC  7lajC  7lakC  7lauC  7layC  7lazC  7lb4C  7lbtC  7lejC  7lekC  7lelC  7lemC  3oniS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| Unit cell |
| |||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 13375.410 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: BRD2, KIAA9001, RING3 / Production host: Homo sapiens (human) / Gene: BRD2, KIAA9001, RING3 / Production host:  | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| #2: Chemical | | #3: Chemical | ChemComp-R78 / | #4: Chemical | ChemComp-CL / | #5: Water | ChemComp-HOH / | Has ligand of interest | Y | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.29 Å3/Da / Density % sol: 46.4 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop Details: 0.2 M Sodium chloride, 0.1 M BIS-TRIS pH 5.5, 25 % w/v Polyethylene glycol 3350 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 22-ID / Wavelength: 1.5418 Å / Beamline: 22-ID / Wavelength: 1.5418 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: DECTRIS EIGER X 16M / Detector: PIXEL / Date: Feb 12, 2019 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.36→35.775 Å / Num. obs: 24096 / % possible obs: 90 % / Redundancy: 3.48 % / Biso Wilson estimate: 11.12 Å2 / CC1/2: 1 / Rmerge(I) obs: 0.022 / Rrim(I) all: 0.025 / Χ2: 1.005 / Net I/σ(I): 32.15 / Num. measured all: 83854 / Scaling rejects: 3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3ONI Resolution: 1.36→35.77 Å / SU ML: 0.1 / Cross valid method: THROUGHOUT / σ(F): 1.36 / Phase error: 16.2 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 67.21 Å2 / Biso mean: 17.2205 Å2 / Biso min: 6.42 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 1.36→35.77 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Rfactor Rfree error: 0 / Total num. of bins used: 8
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj