+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: PDB / ID: 7kc0 | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Structure of the Saccharomyces cerevisiae replicative polymerase delta in complex with a primer/template and the PCNA clamp | |||||||||||||||||||||||||||
 要素 要素 |
| |||||||||||||||||||||||||||
 キーワード キーワード | REPLICATION / Polymerase delta / PCNA / primed DNA / complex | |||||||||||||||||||||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報delta DNA polymerase complex / H3-H4 histone complex chaperone activity / DNA amplification / DNA-templated DNA replication maintenance of fidelity / zeta DNA polymerase complex / RNA-templated DNA biosynthetic process / positive regulation of DNA metabolic process / Mismatch repair (MMR) directed by MSH2:MSH6 (MutSalpha) / meiotic mismatch repair / Processive synthesis on the lagging strand ...delta DNA polymerase complex / H3-H4 histone complex chaperone activity / DNA amplification / DNA-templated DNA replication maintenance of fidelity / zeta DNA polymerase complex / RNA-templated DNA biosynthetic process / positive regulation of DNA metabolic process / Mismatch repair (MMR) directed by MSH2:MSH6 (MutSalpha) / meiotic mismatch repair / Processive synthesis on the lagging strand / Removal of the Flap Intermediate / Polymerase switching / E3 ubiquitin ligases ubiquitinate target proteins / maintenance of DNA trinucleotide repeats / SUMOylation of DNA replication proteins / Translesion synthesis by REV1 / Translesion synthesis by POLK / Translesion synthesis by POLI / 3'-5'-DNA exonuclease activity / nucleotide-excision repair, DNA gap filling / Translesion Synthesis by POLH / establishment of mitotic sister chromatid cohesion / DNA replication proofreading / Termination of translesion DNA synthesis / PCNA complex / DNA replication, removal of RNA primer / lagging strand elongation / double-strand break repair via break-induced replication / postreplication repair / silent mating-type cassette heterochromatin formation / 加水分解酵素; エステル加水分解酵素; 5'-リン酸モノエステル産生エンドデオキシリボヌクレアーゼ / mitotic sister chromatid cohesion / error-free translesion synthesis / DNA metabolic process / DNA strand elongation involved in DNA replication / DNA polymerase processivity factor activity / leading strand elongation / regulation of DNA replication / Dual incision in TC-NER / error-prone translesion synthesis / translesion synthesis / subtelomeric heterochromatin formation / mismatch repair / base-excision repair, gap-filling / positive regulation of DNA repair / positive regulation of DNA replication / replication fork / nucleotide-excision repair / base-excision repair / double-strand break repair via nonhomologous end joining / DNA-templated DNA replication / mitotic cell cycle / 4 iron, 4 sulfur cluster binding / molecular adaptor activity / DNA-directed DNA polymerase / DNA-directed DNA polymerase activity / chromosome, telomeric region / DNA replication / nucleotide binding / DNA binding / zinc ion binding / metal ion binding / identical protein binding / nucleus / cytosol 類似検索 - 分子機能 | |||||||||||||||||||||||||||
| 生物種 |  | |||||||||||||||||||||||||||
| 手法 | 電子顕微鏡法 / 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.2 Å | |||||||||||||||||||||||||||
 データ登録者 データ登録者 | Zheng, F. / Georgescu, R. / Li, H. / O'Donnell, M.E. | |||||||||||||||||||||||||||
| 資金援助 |  米国, 3件 米国, 3件
| |||||||||||||||||||||||||||
 引用 引用 |  ジャーナル: Proc Natl Acad Sci U S A / 年: 2020 ジャーナル: Proc Natl Acad Sci U S A / 年: 2020タイトル: Structure of eukaryotic DNA polymerase δ bound to the PCNA clamp while encircling DNA. 著者: Fengwei Zheng / Roxana E Georgescu / Huilin Li / Michael E O'Donnell /  要旨: The DNA polymerase (Pol) δ of (S.c.) is composed of the catalytic subunit Pol3 along with two regulatory subunits, Pol31 and Pol32. Pol δ binds to proliferating cell nuclear antigen (PCNA) and ...The DNA polymerase (Pol) δ of (S.c.) is composed of the catalytic subunit Pol3 along with two regulatory subunits, Pol31 and Pol32. Pol δ binds to proliferating cell nuclear antigen (PCNA) and functions in genome replication, repair, and recombination. Unique among DNA polymerases, the Pol3 catalytic subunit contains a 4Fe-4S cluster that may sense the cellular redox state. Here we report the 3.2-Å cryo-EM structure of S.c. Pol δ in complex with primed DNA, an incoming ddTTP, and the PCNA clamp. Unexpectedly, Pol δ binds only one subunit of the PCNA trimer. This singular yet extensive interaction holds DNA such that the 2-nm-wide DNA threads through the center of the 3-nm interior channel of the clamp without directly contacting the protein. Thus, a water-mediated clamp and DNA interface enables the PCNA clamp to "waterskate" along the duplex with minimum drag. Pol31 and Pol32 are positioned off to the side of the catalytic Pol3-PCNA-DNA axis. We show here that Pol31-Pol32 binds single-stranded DNA that we propose underlies polymerase recycling during lagging strand synthesis, in analogy to replicase. Interestingly, the 4Fe-4S cluster in the C-terminal CysB domain of Pol3 forms the central interface to Pol31-Pol32, and this strategic location may explain the regulation of the oxidation state on Pol δ activity, possibly useful during cellular oxidative stress. Importantly, human cancer and other disease mutations map to nearly every domain of Pol3, suggesting that all aspects of Pol δ replication are important to human health and disease. | |||||||||||||||||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | 分子:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- ダウンロードとリンク
ダウンロードとリンク
- ダウンロード
ダウンロード
| PDBx/mmCIF形式 |  7kc0.cif.gz 7kc0.cif.gz | 453.2 KB | 表示 |  PDBx/mmCIF形式 PDBx/mmCIF形式 |
|---|---|---|---|---|
| PDB形式 |  pdb7kc0.ent.gz pdb7kc0.ent.gz | 354.7 KB | 表示 |  PDB形式 PDB形式 |
| PDBx/mmJSON形式 |  7kc0.json.gz 7kc0.json.gz | ツリー表示 |  PDBx/mmJSON形式 PDBx/mmJSON形式 | |
| その他 |  その他のダウンロード その他のダウンロード |
-検証レポート
| 文書・要旨 |  7kc0_validation.pdf.gz 7kc0_validation.pdf.gz | 1 MB | 表示 |  wwPDB検証レポート wwPDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  7kc0_full_validation.pdf.gz 7kc0_full_validation.pdf.gz | 1.1 MB | 表示 | |
| XML形式データ |  7kc0_validation.xml.gz 7kc0_validation.xml.gz | 66.6 KB | 表示 | |
| CIF形式データ |  7kc0_validation.cif.gz 7kc0_validation.cif.gz | 103.5 KB | 表示 | |
| アーカイブディレクトリ |  https://data.pdbj.org/pub/pdb/validation_reports/kc/7kc0 https://data.pdbj.org/pub/pdb/validation_reports/kc/7kc0 ftp://data.pdbj.org/pub/pdb/validation_reports/kc/7kc0 ftp://data.pdbj.org/pub/pdb/validation_reports/kc/7kc0 | HTTPS FTP |
-関連構造データ
- リンク
リンク
- 集合体
集合体
| 登録構造単位 | 
|
|---|---|
| 1 |
|
- 要素
要素
-DNA鎖 , 2種, 2分子 PT
| #1: DNA鎖 | 分子量: 7681.985 Da / 分子数: 1 / 由来タイプ: 合成 / 由来: (合成)  |
|---|---|
| #2: DNA鎖 | 分子量: 11677.539 Da / 分子数: 1 / 由来タイプ: 合成 / 由来: (合成)  |
-タンパク質 , 4種, 6分子 FEGABC
| #3: タンパク質 | 分子量: 28944.051 Da / 分子数: 3 / 由来タイプ: 組換発現 由来: (組換発現)  遺伝子: PCNA, POL30, GI526_G0000296, PACBIOSEQ_LOCUS349, PACBIOSEQ_LOCUS352, PACBIOSEQ_LOCUS359, PACBIOSEQ_LOCUS364, PACBIOSEQ_LOCUS365, PACBIOSEQ_LOCUS366, PACBIOSEQ_LOCUS371 発現宿主:  #4: タンパク質 | | 分子量: 124707.359 Da / 分子数: 1 / 由来タイプ: 組換発現 由来: (組換発現)  遺伝子: POL3, GI526_G0000818 / 発現宿主:  参照: UniProt: A0A6A5Q0V0, UniProt: P15436*PLUS, DNA-directed DNA polymerase #5: タンパク質 | | 分子量: 55352.688 Da / 分子数: 1 / 由来タイプ: 組換発現 由来: (組換発現)  遺伝子: POL31, GI526_G0003241 / 発現宿主:  #6: タンパク質 | | 分子量: 40377.715 Da / 分子数: 1 / 由来タイプ: 組換発現 由来: (組換発現)  遺伝子: POL32, GI526_G0003272, PACBIOSEQ_LOCUS3556, PACBIOSEQ_LOCUS3579 発現宿主:  |
|---|
-非ポリマー , 4種, 5分子 

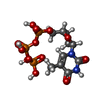




| #7: 化合物 | | #8: 化合物 | ChemComp-SF4 / | #9: 化合物 | ChemComp-D3T / | #10: 化合物 | ChemComp-ZN / | |
|---|
-詳細
| 研究の焦点であるリガンドがあるか | Y |
|---|---|
| Has protein modification | Y |
-実験情報
-実験
| 実験 | 手法: 電子顕微鏡法 |
|---|---|
| EM実験 | 試料の集合状態: PARTICLE / 3次元再構成法: 単粒子再構成法 |
- 試料調製
試料調製
| 構成要素 |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 由来(天然) |
| ||||||||||||||||||||||||
| 由来(組換発現) | 生物種:  | ||||||||||||||||||||||||
| 緩衝液 | pH: 7.5 | ||||||||||||||||||||||||
| 試料 | 包埋: NO / シャドウイング: NO / 染色: NO / 凍結: YES | ||||||||||||||||||||||||
| 急速凍結 | 凍結剤: ETHANE |
- 電子顕微鏡撮影
電子顕微鏡撮影
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
|---|---|
| 顕微鏡 | モデル: FEI TITAN KRIOS |
| 電子銃 | 電子線源:  FIELD EMISSION GUN / 加速電圧: 300 kV / 照射モード: FLOOD BEAM FIELD EMISSION GUN / 加速電圧: 300 kV / 照射モード: FLOOD BEAM |
| 電子レンズ | モード: BRIGHT FIELD |
| 撮影 | 電子線照射量: 68 e/Å2 / フィルム・検出器のモデル: GATAN K3 (6k x 4k) |
- 解析
解析
| ソフトウェア | 名称: PHENIX / バージョン: 1.17.1_3660: / 分類: 精密化 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EMソフトウェア | 名称: PHENIX / カテゴリ: モデル精密化 | ||||||||||||||||||||||||
| CTF補正 | タイプ: NONE | ||||||||||||||||||||||||
| 3次元再構成 | 解像度: 3.2 Å / 解像度の算出法: FSC 0.143 CUT-OFF / 粒子像の数: 133468 / 対称性のタイプ: POINT | ||||||||||||||||||||||||
| 拘束条件 |
|
 ムービー
ムービー コントローラー
コントローラー











 PDBj
PDBj













































