[English] 日本語
 Yorodumi
Yorodumi- EMDB-22803: Cryo-EM 3D map of the Saccharomyces cerevisiae replicative polyme... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22803 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
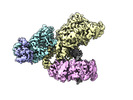
| Title | Cryo-EM 3D map of the Saccharomyces cerevisiae replicative polymerase delta in complex with a primer/template and the PCNA clamp | ||||||||||||
 Map data Map data | Replicative polymerase delta in complex with a primer/template and the PCNA clamp | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Polymerase delta / PCNA / primed DNA / complex / REPLICATION | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationdelta DNA polymerase complex / H3-H4 histone complex chaperone activity / DNA amplification / DNA-templated DNA replication maintenance of fidelity / zeta DNA polymerase complex / Mismatch repair (MMR) directed by MSH2:MSH6 (MutSalpha) / positive regulation of DNA metabolic process / meiotic mismatch repair / RNA-templated DNA biosynthetic process / Processive synthesis on the lagging strand ...delta DNA polymerase complex / H3-H4 histone complex chaperone activity / DNA amplification / DNA-templated DNA replication maintenance of fidelity / zeta DNA polymerase complex / Mismatch repair (MMR) directed by MSH2:MSH6 (MutSalpha) / positive regulation of DNA metabolic process / meiotic mismatch repair / RNA-templated DNA biosynthetic process / Processive synthesis on the lagging strand / Removal of the Flap Intermediate / : / Polymerase switching / maintenance of DNA trinucleotide repeats / SUMOylation of DNA replication proteins / nucleotide-excision repair, DNA gap filling / Translesion synthesis by REV1 / : / : / : / 3'-5'-DNA exonuclease activity / establishment of mitotic sister chromatid cohesion / DNA replication proofreading / : / DNA replication, removal of RNA primer / PCNA complex / lagging strand elongation / double-strand break repair via break-induced replication / DNA damage tolerance / silent mating-type cassette heterochromatin formation / Hydrolases; Acting on ester bonds; Exodeoxyribonucleases producing 5'-phosphomonoesters / mitotic sister chromatid cohesion / error-free translesion synthesis / DNA strand elongation involved in DNA replication / DNA metabolic process / DNA polymerase processivity factor activity / leading strand elongation / regulation of DNA replication / Dual incision in TC-NER / error-prone translesion synthesis / subtelomeric heterochromatin formation / mismatch repair / translesion synthesis / base-excision repair, gap-filling / positive regulation of DNA replication / replication fork / positive regulation of DNA repair / nucleotide-excision repair / base-excision repair / double-strand break repair via nonhomologous end joining / DNA-templated DNA replication / mitotic cell cycle / 4 iron, 4 sulfur cluster binding / DNA-directed DNA polymerase / molecular adaptor activity / DNA-directed DNA polymerase activity / DNA replication / chromosome, telomeric region / nucleotide binding / DNA binding / zinc ion binding / metal ion binding / identical protein binding / nucleus / cytosol Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | ||||||||||||
 Authors Authors | Zheng F / Georgescu R | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2020 Journal: Proc Natl Acad Sci U S A / Year: 2020Title: Structure of eukaryotic DNA polymerase δ bound to the PCNA clamp while encircling DNA. Authors: Fengwei Zheng / Roxana E Georgescu / Huilin Li / Michael E O'Donnell /  Abstract: The DNA polymerase (Pol) δ of (S.c.) is composed of the catalytic subunit Pol3 along with two regulatory subunits, Pol31 and Pol32. Pol δ binds to proliferating cell nuclear antigen (PCNA) and ...The DNA polymerase (Pol) δ of (S.c.) is composed of the catalytic subunit Pol3 along with two regulatory subunits, Pol31 and Pol32. Pol δ binds to proliferating cell nuclear antigen (PCNA) and functions in genome replication, repair, and recombination. Unique among DNA polymerases, the Pol3 catalytic subunit contains a 4Fe-4S cluster that may sense the cellular redox state. Here we report the 3.2-Å cryo-EM structure of S.c. Pol δ in complex with primed DNA, an incoming ddTTP, and the PCNA clamp. Unexpectedly, Pol δ binds only one subunit of the PCNA trimer. This singular yet extensive interaction holds DNA such that the 2-nm-wide DNA threads through the center of the 3-nm interior channel of the clamp without directly contacting the protein. Thus, a water-mediated clamp and DNA interface enables the PCNA clamp to "waterskate" along the duplex with minimum drag. Pol31 and Pol32 are positioned off to the side of the catalytic Pol3-PCNA-DNA axis. We show here that Pol31-Pol32 binds single-stranded DNA that we propose underlies polymerase recycling during lagging strand synthesis, in analogy to replicase. Interestingly, the 4Fe-4S cluster in the C-terminal CysB domain of Pol3 forms the central interface to Pol31-Pol32, and this strategic location may explain the regulation of the oxidation state on Pol δ activity, possibly useful during cellular oxidative stress. Importantly, human cancer and other disease mutations map to nearly every domain of Pol3, suggesting that all aspects of Pol δ replication are important to human health and disease. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22803.map.gz emd_22803.map.gz | 267.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22803-v30.xml emd-22803-v30.xml emd-22803.xml emd-22803.xml | 22.1 KB 22.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_22803.png emd_22803.png | 157.6 KB | ||
| Filedesc metadata |  emd-22803.cif.gz emd-22803.cif.gz | 7.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22803 http://ftp.pdbj.org/pub/emdb/structures/EMD-22803 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22803 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22803 | HTTPS FTP |
-Related structure data
| Related structure data |  7kc0MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22803.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22803.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Replicative polymerase delta in complex with a primer/template and the PCNA clamp | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.826 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Pol delta-PCNA-DNA
+Supramolecule #1: Pol delta-PCNA-DNA
+Supramolecule #2: dsDNA
+Supramolecule #3: Proteins
+Macromolecule #1: DNA (5'-D(P*AP*TP*GP*AP*CP*CP*AP*TP*GP*AP*TP*TP*AP*CP*GP*AP*AP*TP...
+Macromolecule #2: DNA (25-MER)
+Macromolecule #3: Proliferating cell nuclear antigen
+Macromolecule #4: DNA polymerase
+Macromolecule #5: POL31 isoform 1
+Macromolecule #6: POL32 isoform 1
+Macromolecule #7: MAGNESIUM ION
+Macromolecule #8: IRON/SULFUR CLUSTER
+Macromolecule #9: 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE
+Macromolecule #10: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 68.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)