[English] 日本語
 Yorodumi
Yorodumi- PDB-7joz: Crystal structure of dopamine D1 receptor in complex with G prote... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7joz | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of dopamine D1 receptor in complex with G protein and a non-catechol agonist | ||||||
 Components Components |
| ||||||
 Keywords Keywords | MEMBRANE PROTEIN / Dopamine receptor / D1 / non-catechol agonist / G protein | ||||||
| Function / homology |  Function and homology information Function and homology informationdopamine neurotransmitter receptor activity, coupled via Gs / dopamine neurotransmitter receptor activity / cerebral cortex GABAergic interneuron migration / Dopamine receptors / regulation of dopamine uptake involved in synaptic transmission / dopamine binding / operant conditioning / phospholipase C-activating dopamine receptor signaling pathway / peristalsis / heterotrimeric G-protein binding ...dopamine neurotransmitter receptor activity, coupled via Gs / dopamine neurotransmitter receptor activity / cerebral cortex GABAergic interneuron migration / Dopamine receptors / regulation of dopamine uptake involved in synaptic transmission / dopamine binding / operant conditioning / phospholipase C-activating dopamine receptor signaling pathway / peristalsis / heterotrimeric G-protein binding / modification of postsynaptic structure / G protein-coupled receptor complex / positive regulation of neuron migration / habituation / grooming behavior / regulation of dopamine metabolic process / dopamine transport / astrocyte development / dentate gyrus development / sensitization / striatum development / positive regulation of potassium ion transport / conditioned taste aversion / maternal behavior / arrestin family protein binding / non-motile cilium / G protein-coupled dopamine receptor signaling pathway / long-term synaptic depression / adult walking behavior / mating behavior / : / ciliary membrane / temperature homeostasis / dopamine metabolic process / transmission of nerve impulse / PKA activation in glucagon signalling / developmental growth / hair follicle placode formation / G-protein alpha-subunit binding / intracellular transport / D1 dopamine receptor binding / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / viral release from host cell by cytolysis / positive regulation of synaptic transmission, glutamatergic / vascular endothelial cell response to laminar fluid shear stress / renal water homeostasis / activation of adenylate cyclase activity / behavioral fear response / Hedgehog 'off' state / neuronal action potential / adenylate cyclase-activating adrenergic receptor signaling pathway / behavioral response to cocaine / synapse assembly / peptidoglycan catabolic process / regulation of insulin secretion / cellular response to glucagon stimulus / presynaptic modulation of chemical synaptic transmission / positive regulation of release of sequestered calcium ion into cytosol / adenylate cyclase activator activity / trans-Golgi network membrane / response to amphetamine / synaptic transmission, glutamatergic / negative regulation of inflammatory response to antigenic stimulus / bone development / visual learning / G protein-coupled receptor activity / platelet aggregation / vasodilation / GABA-ergic synapse / cognition / memory / G-protein beta/gamma-subunit complex binding / adenylate cyclase-activating G protein-coupled receptor signaling pathway / protein import into nucleus / Olfactory Signaling Pathway / cell wall macromolecule catabolic process / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / lysozyme / G beta:gamma signalling through BTK / lysozyme activity / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / Sensory perception of sweet, bitter, and umami (glutamate) taste / sensory perception of smell / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human)  Enterobacteria phage T4 (virus) Enterobacteria phage T4 (virus) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.8 Å MOLECULAR REPLACEMENT / Resolution: 3.8 Å | ||||||
 Authors Authors | Sun, B. / Feng, D. / Chu, M.L. / Fish, I. / Kelm, S. / Lebon, F. / Lovera, S. / Valade, A. / Wood, M. / Ceska, T. ...Sun, B. / Feng, D. / Chu, M.L. / Fish, I. / Kelm, S. / Lebon, F. / Lovera, S. / Valade, A. / Wood, M. / Ceska, T. / Kobilka, T.S. / Sands, Z. / Kobilka, B.K. | ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Crystal structure of dopamine D1 receptor in complex with G protein and a non-catechol agonist. Authors: Sun, B. / Feng, D. / Chu, M.L. / Fish, I. / Lovera, S. / Sands, Z.A. / Kelm, S. / Valade, A. / Wood, M. / Ceska, T. / Kobilka, T.S. / Lebon, F. / Kobilka, B.K. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7joz.cif.gz 7joz.cif.gz | 557.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7joz.ent.gz pdb7joz.ent.gz | 400.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7joz.json.gz 7joz.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/jo/7joz https://data.pdbj.org/pub/pdb/validation_reports/jo/7joz ftp://data.pdbj.org/pub/pdb/validation_reports/jo/7joz ftp://data.pdbj.org/pub/pdb/validation_reports/jo/7joz | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3sn6S S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||
| Unit cell |
|
- Components
Components
-Guanine nucleotide-binding protein ... , 3 types, 3 molecules ABG
| #1: Protein | Mass: 44326.160 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GNAS, GNAS1, GSP / Production host: Homo sapiens (human) / Gene: GNAS, GNAS1, GSP / Production host:  Trichoplusia ni (cabbage looper) / References: UniProt: P63092 Trichoplusia ni (cabbage looper) / References: UniProt: P63092 |
|---|---|
| #2: Protein | Mass: 38534.062 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GNB1 / Production host: Homo sapiens (human) / Gene: GNB1 / Production host:  Trichoplusia ni (cabbage looper) / References: UniProt: P62873 Trichoplusia ni (cabbage looper) / References: UniProt: P62873 |
| #3: Protein | Mass: 7861.143 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GNG2 / Production host: Homo sapiens (human) / Gene: GNG2 / Production host:  Trichoplusia ni (cabbage looper) / References: UniProt: P59768 Trichoplusia ni (cabbage looper) / References: UniProt: P59768 |
-Antibody / Protein , 2 types, 2 molecules NR
| #4: Antibody | Mass: 17352.498 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| #5: Protein | Mass: 59856.949 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Enterobacteria phage T4 (virus), (gene. exp.) Enterobacteria phage T4 (virus), (gene. exp.)  Homo sapiens (human) Homo sapiens (human)Gene: e, T4Tp126, DRD1 / Production host:  |
-Non-polymers , 2 types, 2 molecules 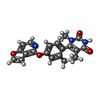


| #6: Chemical | ChemComp-VFP / |
|---|---|
| #7: Chemical | ChemComp-1PE / |
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.13 Å3/Da / Density % sol: 65.6 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: lipidic cubic phase / pH: 5.5 Details: 0.1M MES pH 6.5, 0.1M Ammonium Dibasic Citrate, 18-25% (w/w) PEG400, 1mM TCEP, 10uM UCB1575401 |
-Data collection
| Diffraction | Mean temperature: 80 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 23-ID-D / Wavelength: 1.0332 Å / Beamline: 23-ID-D / Wavelength: 1.0332 Å |
| Detector | Type: DECTRIS PILATUS3 6M / Detector: PIXEL / Date: Oct 1, 2015 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.0332 Å / Relative weight: 1 |
| Reflection | Resolution: 3.79→49 Å / Num. obs: 20110 / % possible obs: 94.3 % / Redundancy: 3.6 % / Biso Wilson estimate: 127.32 Å2 / CC1/2: 0.99 / Rmerge(I) obs: 0.189 / Rpim(I) all: 0.116 / Net I/σ(I): 4.7 |
| Reflection shell | Resolution: 3.79→4.16 Å / Redundancy: 3.7 % / Rmerge(I) obs: 0.904 / Num. unique obs: 4783 / CC1/2: 0.468 / Rpim(I) all: 0.555 / % possible all: 95.2 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3SN6 Resolution: 3.8→47 Å / SU ML: 0.5539 / Cross valid method: FREE R-VALUE / σ(F): 1.33 / Phase error: 31.3177 Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 139.93 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3.8→47 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Origin x: -2.32450069099 Å / Origin y: -38.0969843562 Å / Origin z: -40.5635125984 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group | Selection details: all |
 Movie
Movie Controller
Controller










 PDBj
PDBj


























