[English] 日本語
 Yorodumi
Yorodumi- PDB-7f8u: Crystal structure of the cholecystokinin receptor CCKAR in comple... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7f8u | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the cholecystokinin receptor CCKAR in complex with lintitript | |||||||||||||||||||||||||||||||||
 Components Components | Fusion protein of Cholecystokinin receptor type A and Endolysin | |||||||||||||||||||||||||||||||||
 Keywords Keywords | STRUCTURAL PROTEIN / G protein-coulped receptor / Cholecystokinin receptor CCKAR / lintitript | |||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationcholecystokinin receptor activity / cholecystokinin signaling pathway / regulation of hormone secretion / forebrain development / peptide hormone binding / viral release from host cell by cytolysis / cellular response to hormone stimulus / peptidoglycan catabolic process / axonogenesis / Peptide ligand-binding receptors ...cholecystokinin receptor activity / cholecystokinin signaling pathway / regulation of hormone secretion / forebrain development / peptide hormone binding / viral release from host cell by cytolysis / cellular response to hormone stimulus / peptidoglycan catabolic process / axonogenesis / Peptide ligand-binding receptors / neuron migration / cell wall macromolecule catabolic process / lysozyme / lysozyme activity / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / host cell cytoplasm / defense response to bacterium / G protein-coupled receptor signaling pathway / nucleoplasm / membrane / plasma membrane / cytosol Similarity search - Function | |||||||||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) Enterobacteria phage T4 (virus) Enterobacteria phage T4 (virus) | |||||||||||||||||||||||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.8 Å MOLECULAR REPLACEMENT / Resolution: 2.8 Å | |||||||||||||||||||||||||||||||||
 Authors Authors | Zhang, X. / He, C. / Wang, M. / Zhou, Q. / Yang, D. / Zhu, Y. / Wu, B. / Zhao, Q. | |||||||||||||||||||||||||||||||||
| Funding support |  China, 10items China, 10items
| |||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2021 Journal: Nat Chem Biol / Year: 2021Title: Structures of the human cholecystokinin receptors bound to agonists and antagonists. Authors: Xuefeng Zhang / Chenglin He / Mu Wang / Qingtong Zhou / Dehua Yang / Ya Zhu / Wenbo Feng / Hui Zhang / Antao Dai / Xiaojing Chu / Jia Wang / Zhenlin Yang / Yi Jiang / Ulrich Sensfuss / ...Authors: Xuefeng Zhang / Chenglin He / Mu Wang / Qingtong Zhou / Dehua Yang / Ya Zhu / Wenbo Feng / Hui Zhang / Antao Dai / Xiaojing Chu / Jia Wang / Zhenlin Yang / Yi Jiang / Ulrich Sensfuss / Qiuxiang Tan / Shuo Han / Steffen Reedtz-Runge / H Eric Xu / Suwen Zhao / Ming-Wei Wang / Beili Wu / Qiang Zhao /   Abstract: Cholecystokinin receptors, CCKR and CCKR, are important neurointestinal peptide hormone receptors and play a vital role in food intake and appetite regulation. Here, we report three crystal ...Cholecystokinin receptors, CCKR and CCKR, are important neurointestinal peptide hormone receptors and play a vital role in food intake and appetite regulation. Here, we report three crystal structures of the human CCKR in complex with different ligands, including one peptide agonist and two small-molecule antagonists, as well as two cryo-electron microscopy structures of CCKR-gastrin in complex with G and G, respectively. These structures reveal the recognition pattern of different ligand types and the molecular basis of peptide selectivity in the cholecystokinin receptor family. By comparing receptor structures in different conformational states, a stepwise activation process of cholecystokinin receptors is proposed. Combined with pharmacological data, our results provide atomic details for differential ligand recognition and receptor activation mechanisms. These insights will facilitate the discovery of potential therapeutics targeting cholecystokinin receptors. | |||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7f8u.cif.gz 7f8u.cif.gz | 104.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7f8u.ent.gz pdb7f8u.ent.gz | 76 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7f8u.json.gz 7f8u.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  7f8u_validation.pdf.gz 7f8u_validation.pdf.gz | 731.4 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  7f8u_full_validation.pdf.gz 7f8u_full_validation.pdf.gz | 742.7 KB | Display | |
| Data in XML |  7f8u_validation.xml.gz 7f8u_validation.xml.gz | 18.4 KB | Display | |
| Data in CIF |  7f8u_validation.cif.gz 7f8u_validation.cif.gz | 24.2 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/f8/7f8u https://data.pdbj.org/pub/pdb/validation_reports/f8/7f8u ftp://data.pdbj.org/pub/pdb/validation_reports/f8/7f8u ftp://data.pdbj.org/pub/pdb/validation_reports/f8/7f8u | HTTPS FTP |
-Related structure data
| Related structure data |  7f8vC  7f8wC  7f8xC  7f8yC  5zbqS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 60450.367 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human), (gene. exp.) Homo sapiens (human), (gene. exp.)  Enterobacteria phage T4 (virus) Enterobacteria phage T4 (virus)Gene: CCKAR, CCKRA / Production host:  |
|---|---|
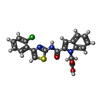
| #2: Chemical | ChemComp-1OE / |
| Has ligand of interest | Y |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.74 Å3/Da / Density % sol: 55.04 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: lipidic cubic phase Details: 0.1 M HEPES, pH7.5, 25% PEG400, 250 mM sodium tartrate ,1% 1,2 -butanediol |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SPring-8 SPring-8  / Beamline: BL41XU / Wavelength: 1 Å / Beamline: BL41XU / Wavelength: 1 Å |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: May 25, 2016 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 2.8→50 Å / Num. obs: 15523 / % possible obs: 97.5 % / Redundancy: 4.1 % / Biso Wilson estimate: 87.7 Å2 / CC1/2: 0.985 / Net I/σ(I): 5.21 |
| Reflection shell | Resolution: 2.8→2.87 Å / Mean I/σ(I) obs: 1 / Num. unique obs: 1545 / CC1/2: 0.77 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5ZBQ Resolution: 2.8→29.63 Å / Cross valid method: FREE R-VALUE Stereochemistry target values: GEOSTD + MONOMER LIBRARY + CDL V1.2
| ||||||||||||||||||||||||
| Displacement parameters | Biso mean: 91.14 Å2 | ||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.8→29.63 Å
| ||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.8→2.9 Å
|
 Movie
Movie Controller
Controller














 PDBj
PDBj