[English] 日本語
 Yorodumi
Yorodumi- EMDB-31494: Cryo-EM structure of the cholecystokinin receptor CCKBR in comple... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31494 | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
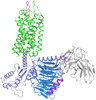
| Title | Cryo-EM structure of the cholecystokinin receptor CCKBR in complex with gastrin-17 and Gq | |||||||||||||||||||||||||||||||||
 Map data Map data | CCKBR-Gq complex | |||||||||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationgastrin receptor activity / type B gastrin/cholecystokinin receptor binding / gland development / cholecystokinin receptor activity / cholecystokinin signaling pathway / pH reduction / gastric acid secretion / sensory perception of itch / Fatty Acids bound to GPR40 (FFAR1) regulate insulin secretion / Acetylcholine regulates insulin secretion ...gastrin receptor activity / type B gastrin/cholecystokinin receptor binding / gland development / cholecystokinin receptor activity / cholecystokinin signaling pathway / pH reduction / gastric acid secretion / sensory perception of itch / Fatty Acids bound to GPR40 (FFAR1) regulate insulin secretion / Acetylcholine regulates insulin secretion / phospholipase C-activating G protein-coupled glutamate receptor signaling pathway / neuropeptide receptor activity / PLC beta mediated events / phospholipase C-activating serotonin receptor signaling pathway / 1-phosphatidylinositol-3-kinase regulator activity / regulation of platelet activation / entrainment of circadian clock / Gastrin-CREB signalling pathway via PKC and MAPK / digestive tract development / regulation of canonical Wnt signaling pathway / cellular response to acidic pH / glutamate receptor signaling pathway / mast cell degranulation / peptide hormone binding / phototransduction, visible light / photoreceptor outer segment / postsynaptic cytosol / neuropeptide signaling pathway / response to prostaglandin E / hormone-mediated signaling pathway / GTPase activator activity / Turbulent (oscillatory, disturbed) flow shear stress activates signaling by PIEZO1 and integrins in endothelial cells / Peptide ligand-binding receptors / G protein-coupled receptor binding / G-protein beta/gamma-subunit complex binding / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Olfactory Signaling Pathway / Activation of the phototransduction cascade / blood coagulation / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / Sensory perception of sweet, bitter, and umami (glutamate) taste / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / ADORA2B mediated anti-inflammatory cytokines production / cellular response to catecholamine stimulus / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / Inactivation, recovery and regulation of the phototransduction cascade / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / positive regulation of cytosolic calcium ion concentration / retina development in camera-type eye / G protein activity / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / nuclear membrane / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Ras protein signal transduction / cell surface receptor signaling pathway / Extra-nuclear estrogen signaling / cell population proliferation / protein stabilization / G protein-coupled receptor signaling pathway / intracellular membrane-bounded organelle / lysosomal membrane / GTPase activity / positive regulation of cell population proliferation / synapse / GTP binding Similarity search - Function | |||||||||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||||||||||||||||||||||||||
 Authors Authors | Zhang X / He C / Wang M / Zhou Q / Yang D / Zhu Y / Wu B / Zhao Q | |||||||||||||||||||||||||||||||||
| Funding support |  China, 10 items China, 10 items
| |||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2021 Journal: Nat Chem Biol / Year: 2021Title: Structures of the human cholecystokinin receptors bound to agonists and antagonists. Authors: Xuefeng Zhang / Chenglin He / Mu Wang / Qingtong Zhou / Dehua Yang / Ya Zhu / Wenbo Feng / Hui Zhang / Antao Dai / Xiaojing Chu / Jia Wang / Zhenlin Yang / Yi Jiang / Ulrich Sensfuss / ...Authors: Xuefeng Zhang / Chenglin He / Mu Wang / Qingtong Zhou / Dehua Yang / Ya Zhu / Wenbo Feng / Hui Zhang / Antao Dai / Xiaojing Chu / Jia Wang / Zhenlin Yang / Yi Jiang / Ulrich Sensfuss / Qiuxiang Tan / Shuo Han / Steffen Reedtz-Runge / H Eric Xu / Suwen Zhao / Ming-Wei Wang / Beili Wu / Qiang Zhao /   Abstract: Cholecystokinin receptors, CCKR and CCKR, are important neurointestinal peptide hormone receptors and play a vital role in food intake and appetite regulation. Here, we report three crystal ...Cholecystokinin receptors, CCKR and CCKR, are important neurointestinal peptide hormone receptors and play a vital role in food intake and appetite regulation. Here, we report three crystal structures of the human CCKR in complex with different ligands, including one peptide agonist and two small-molecule antagonists, as well as two cryo-electron microscopy structures of CCKR-gastrin in complex with G and G, respectively. These structures reveal the recognition pattern of different ligand types and the molecular basis of peptide selectivity in the cholecystokinin receptor family. By comparing receptor structures in different conformational states, a stepwise activation process of cholecystokinin receptors is proposed. Combined with pharmacological data, our results provide atomic details for differential ligand recognition and receptor activation mechanisms. These insights will facilitate the discovery of potential therapeutics targeting cholecystokinin receptors. | |||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31494.map.gz emd_31494.map.gz | 5.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31494-v30.xml emd-31494-v30.xml emd-31494.xml emd-31494.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31494.png emd_31494.png | 116.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31494 http://ftp.pdbj.org/pub/emdb/structures/EMD-31494 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31494 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31494 | HTTPS FTP |
-Related structure data
| Related structure data |  7f8wMC  7f8uC  7f8vC  7f8xC  7f8yC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31494.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31494.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CCKBR-Gq complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.045 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Cholecystokinin recetor CCKBR in complex with gastrin-17 and Gq
+Supramolecule #1: Cholecystokinin recetor CCKBR in complex with gastrin-17 and Gq
+Supramolecule #2: Guanine nucleotide-binding protein G subunits
+Supramolecule #3: scFv16
+Supramolecule #4: Gastrin-17
+Supramolecule #5: cholecystokinin type B receptor
+Macromolecule #1: Guanine nucleotide-binding protein G(q) subunit alpha
+Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
+Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
+Macromolecule #4: scFv16
+Macromolecule #5: Gastrin-17
+Macromolecule #6: Gastrin/cholecystokinin type B receptor
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 1.75 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 354647 |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller





































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















 Baculovirus expression vector pFastBac1-HM
Baculovirus expression vector pFastBac1-HM