[English] 日本語
 Yorodumi
Yorodumi- EMDB-30603: Cryo-EM structure of the cortisol-bound adhesion receptor GPR97-G... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30603 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the cortisol-bound adhesion receptor GPR97-Go complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / GPR97 / complex / adhesion G protein-coupled receptor / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information: / mu-type opioid receptor binding / corticotropin-releasing hormone receptor 1 binding / negative regulation of non-canonical NF-kappaB signal transduction / vesicle docking involved in exocytosis / G protein-coupled dopamine receptor signaling pathway / regulation of heart contraction / parallel fiber to Purkinje cell synapse / postsynaptic modulation of chemical synaptic transmission / specific granule membrane ...: / mu-type opioid receptor binding / corticotropin-releasing hormone receptor 1 binding / negative regulation of non-canonical NF-kappaB signal transduction / vesicle docking involved in exocytosis / G protein-coupled dopamine receptor signaling pathway / regulation of heart contraction / parallel fiber to Purkinje cell synapse / postsynaptic modulation of chemical synaptic transmission / specific granule membrane / G protein-coupled serotonin receptor binding / adenylate cyclase regulator activity / adenylate cyclase-inhibiting serotonin receptor signaling pathway / regulation of cell migration / B cell differentiation / muscle contraction / locomotory behavior / negative regulation of insulin secretion / G protein-coupled receptor activity / GABA-ergic synapse / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / G alpha (12/13) signalling events / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / presynaptic membrane / G protein activity / retina development in camera-type eye / cell body / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / postsynaptic membrane / Ras protein signal transduction / cell surface receptor signaling pathway / Extra-nuclear estrogen signaling / cell population proliferation / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / dendrite / synapse / Neutrophil degranulation / GTP binding / protein-containing complex binding / glutamatergic synapse / signal transduction / extracellular exosome / metal ion binding / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
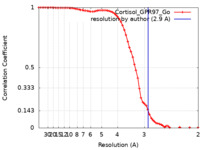
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Ping Y / Mao C | |||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Structures of the glucocorticoid-bound adhesion receptor GPR97-G complex. Authors: Yu-Qi Ping / Chunyou Mao / Peng Xiao / Ru-Jia Zhao / Yi Jiang / Zhao Yang / Wen-Tao An / Dan-Dan Shen / Fan Yang / Huibing Zhang / Changxiu Qu / Qingya Shen / Caiping Tian / Zi-Jian Li / ...Authors: Yu-Qi Ping / Chunyou Mao / Peng Xiao / Ru-Jia Zhao / Yi Jiang / Zhao Yang / Wen-Tao An / Dan-Dan Shen / Fan Yang / Huibing Zhang / Changxiu Qu / Qingya Shen / Caiping Tian / Zi-Jian Li / Shaolong Li / Guang-Yu Wang / Xiaona Tao / Xin Wen / Ya-Ni Zhong / Jing Yang / Fan Yi / Xiao Yu / H Eric Xu / Yan Zhang / Jin-Peng Sun /  Abstract: Adhesion G-protein-coupled receptors (GPCRs) are a major family of GPCRs, but limited knowledge of their ligand regulation or structure is available. Here we report that glucocorticoid stress ...Adhesion G-protein-coupled receptors (GPCRs) are a major family of GPCRs, but limited knowledge of their ligand regulation or structure is available. Here we report that glucocorticoid stress hormones activate adhesion G-protein-coupled receptor G3 (ADGRG3; also known as GPR97), a prototypical adhesion GPCR. The cryo-electron microscopy structures of GPR97-G complexes bound to the anti-inflammatory drug beclomethasone or the steroid hormone cortisol revealed that glucocorticoids bind to a pocket within the transmembrane domain. The steroidal core of glucocorticoids is packed against the 'toggle switch' residue W, which senses the binding of a ligand and induces activation of the receptor. Active GPR97 uses a quaternary core and HLY motif to fasten the seven-transmembrane bundle and to mediate G protein coupling. The cytoplasmic side of GPR97 has an open cavity, where all three intracellular loops interact with the G protein, contributing to the high basal activity of GRP97. Palmitoylation at the cytosolic tail of the G protein was found to be essential for efficient engagement with GPR97 but is not observed in other solved GPCR complex structures. Our work provides a structural basis for ligand binding to the seven-transmembrane domain of an adhesion GPCR and subsequent G protein coupling. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30603.map.gz emd_30603.map.gz | 18 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30603-v30.xml emd-30603-v30.xml emd-30603.xml emd-30603.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_30603_fsc.xml emd_30603_fsc.xml | 6.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_30603.png emd_30603.png | 61.9 KB | ||
| Filedesc metadata |  emd-30603.cif.gz emd-30603.cif.gz | 7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30603 http://ftp.pdbj.org/pub/emdb/structures/EMD-30603 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30603 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30603 | HTTPS FTP |
-Related structure data
| Related structure data |  7d77MC  7d76C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30603.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30603.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.014 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
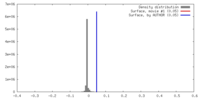
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Cortisol-bound adhesion receptor GPR97-Go complex
+Supramolecule #1: Cortisol-bound adhesion receptor GPR97-Go complex
+Supramolecule #2: Adhesion G protein-coupled receptor G3; GPR97
+Supramolecule #3: Go
+Supramolecule #4: scFv16
+Macromolecule #1: Guanine nucleotide-binding protein G(o) subunit alpha
+Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
+Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
+Macromolecule #4: Adhesion G protein-coupled receptor G3; GPR97
+Macromolecule #5: scFv16
+Macromolecule #6: PALMITIC ACID
+Macromolecule #7: CHOLESTEROL HEMISUCCINATE
+Macromolecule #8: (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione
+Macromolecule #9: CHOLESTEROL
+Macromolecule #10: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 20 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil / Material: GOLD / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 5871 / Average electron dose: 62.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 49310 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)