+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6zfx | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | hSARM1 GraFix-ed | |||||||||
 Components Components | NAD(+) hydrolase SARM1 | |||||||||
 Keywords Keywords | HYDROLASE / NADase / ARM domain / SAM domain / TIR domain | |||||||||
| Function / homology |  Function and homology information Function and homology informationextrinsic component of synaptic membrane / negative regulation of MyD88-independent toll-like receptor signaling pathway / MyD88-independent TLR4 cascade / NADP+ nucleosidase activity / Toll Like Receptor 3 (TLR3) Cascade / NAD+ catabolic process / NAD+ nucleosidase activity / regulation of synapse pruning / modification of postsynaptic structure / ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase ...extrinsic component of synaptic membrane / negative regulation of MyD88-independent toll-like receptor signaling pathway / MyD88-independent TLR4 cascade / NADP+ nucleosidase activity / Toll Like Receptor 3 (TLR3) Cascade / NAD+ catabolic process / NAD+ nucleosidase activity / regulation of synapse pruning / modification of postsynaptic structure / ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase / protein localization to mitochondrion / NAD+ nucleosidase activity, cyclic ADP-ribose generating / nervous system process / Hydrolases; Glycosylases; Hydrolysing N-glycosyl compounds / regulation of dendrite morphogenesis / response to glucose / response to axon injury / regulation of neuron apoptotic process / signaling adaptor activity / TRAF6-mediated induction of TAK1 complex within TLR4 complex / Activation of IRF3, IRF7 mediated by TBK1, IKKε (IKBKE) / IKK complex recruitment mediated by RIP1 / neuromuscular junction / nervous system development / microtubule / mitochondrial outer membrane / cell differentiation / axon / innate immune response / synapse / dendrite / glutamatergic synapse / cell surface / signal transduction / protein-containing complex / mitochondrion / identical protein binding / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.88 Å | |||||||||
 Authors Authors | Sporny, M. / Guez-Haddad, J. / Khazma, T. / Yaron, A. / Dessau, M. / Mim, C. / Isupov, M.N. / Zalk, R. / Hons, M. / Opatowsky, Y. | |||||||||
| Funding support |  Israel, 2items Israel, 2items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: Structural basis for SARM1 inhibition and activation under energetic stress. Authors: Michael Sporny / Julia Guez-Haddad / Tami Khazma / Avraham Yaron / Moshe Dessau / Yoel Shkolnisky / Carsten Mim / Michail N Isupov / Ran Zalk / Michael Hons / Yarden Opatowsky /     Abstract: SARM1, an executor of axonal degeneration, displays NADase activity that depletes the key cellular metabolite, NAD+, in response to nerve injury. The basis of SARM1 inhibition and its activation ...SARM1, an executor of axonal degeneration, displays NADase activity that depletes the key cellular metabolite, NAD+, in response to nerve injury. The basis of SARM1 inhibition and its activation under stress conditions are still unknown. Here, we present cryo-EM maps of SARM1 at 2.9 and 2.7 Å resolutions. These indicate that SARM1 homo-octamer avoids premature activation by assuming a packed conformation, with ordered inner and peripheral rings, that prevents dimerization and activation of the catalytic domains. This inactive conformation is stabilized by binding of SARM1's own substrate NAD+ in an allosteric location, away from the catalytic sites. This model was validated by mutagenesis of the allosteric site, which led to constitutively active SARM1. We propose that the reduction of cellular NAD+ concentration contributes to the disassembly of SARM1's peripheral ring, which allows formation of active NADase domain dimers, thereby further depleting NAD+ to cause an energetic catastrophe and cell death. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6zfx.cif.gz 6zfx.cif.gz | 970.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6zfx.ent.gz pdb6zfx.ent.gz | 811.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6zfx.json.gz 6zfx.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/zf/6zfx https://data.pdbj.org/pub/pdb/validation_reports/zf/6zfx ftp://data.pdbj.org/pub/pdb/validation_reports/zf/6zfx ftp://data.pdbj.org/pub/pdb/validation_reports/zf/6zfx | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  11187MC  6zg0C  6zg1C  7anwC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments: Component-ID: 1 / Beg auth comp-ID: GLY / Beg label comp-ID: GLY / End auth comp-ID: GLN / End label comp-ID: GLN / Refine code: 1 / Auth seq-ID: 56 - 700 / Label seq-ID: 58 - 702
|
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera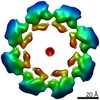









 PDBj
PDBj


