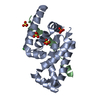
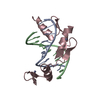
Entry Database : PDB / ID : 6xqiTitle Structure of HIV-1 Vpr in complex with the human nucleotide excision repair protein hHR23A ASN-PRO-LEU-GLU-PHE-LEU Protein Vpr UV excision repair protein RAD23 homolog A Keywords / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / / / Resolution : 2.34 Å Authors Calero, G.C. / Wu, Y. / Weiss, S.C. Funding support Organization Grant number Country National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) P50AI150481
Journal : Nat Commun / Year : 2021Title : Structure of HIV-1 Vpr in complex with the human nucleotide excision repair protein hHR23A.Authors : Byeon, I.L. / Calero, G. / Wu, Y. / Byeon, C.H. / Jung, J. / DeLucia, M. / Zhou, X. / Weiss, S. / Ahn, J. / Hao, C. / Skowronski, J. / Gronenborn, A.M. History Deposition Jul 9, 2020 Deposition site / Processing site Revision 1.0 Aug 11, 2021 Provider / Type Revision 1.1 Dec 8, 2021 Group / Category / citation_authorItem _citation.country / _citation.journal_abbrev ... _citation.country / _citation.journal_abbrev / _citation.journal_id_CSD / _citation.journal_id_ISSN / _citation.journal_volume / _citation.page_first / _citation.page_last / _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation.year / _citation_author.identifier_ORCID / _citation_author.name Revision 1.2 May 22, 2024 Group / Category / chem_comp_bond
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Human immunodeficiency virus type 1 group M subtype B
Human immunodeficiency virus type 1 group M subtype B Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  SAD / Resolution: 2.34 Å
SAD / Resolution: 2.34 Å  Authors
Authors United States, 1items
United States, 1items  Citation
Citation Journal: Nat Commun / Year: 2021
Journal: Nat Commun / Year: 2021 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6xqi.cif.gz
6xqi.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6xqi.ent.gz
pdb6xqi.ent.gz PDB format
PDB format 6xqi.json.gz
6xqi.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/xq/6xqi
https://data.pdbj.org/pub/pdb/validation_reports/xq/6xqi ftp://data.pdbj.org/pub/pdb/validation_reports/xq/6xqi
ftp://data.pdbj.org/pub/pdb/validation_reports/xq/6xqi Links
Links Assembly
Assembly


 Movie
Movie Controller
Controller










 PDBj
PDBj



