[English] 日本語
 Yorodumi
Yorodumi- PDB-6vtq: Crystal structure of G16C human Galectin-7 mutant in complex with... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6vtq | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of G16C human Galectin-7 mutant in complex with lactose | |||||||||||||||||||||
 Components Components | Galectin-7 | |||||||||||||||||||||
 Keywords Keywords | SUGAR BINDING PROTEIN / Human Galectin-7 / lactose / G16C mutant / disulfide bond | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationDifferentiation of Keratinocytes in Interfollicular Epidermis in Mammalian Skin / heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules / carbohydrate binding / apoptotic process / extracellular space / extracellular exosome / nucleus / cytoplasm Similarity search - Function | |||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.95 Å molecular replacement / Resolution: 1.95 Å | |||||||||||||||||||||
 Authors Authors | Pham, N.T.H. / Calmettes, C. / Doucet, N. | |||||||||||||||||||||
| Funding support |  Canada, Canada,  United States, 6items United States, 6items
| |||||||||||||||||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2021 Journal: J.Biol.Chem. / Year: 2021Title: Perturbing dimer interactions and allosteric communication modulates the immunosuppressive activity of human galectin-7. Authors: Pham, N.T.H. / Letourneau, M. / Fortier, M. / Begin, G. / Al-Abdul-Wahid, M.S. / Pucci, F. / Folch, B. / Rooman, M. / Chatenet, D. / St-Pierre, Y. / Lague, P. / Calmettes, C. / Doucet, N. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6vtq.cif.gz 6vtq.cif.gz | 117.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6vtq.ent.gz pdb6vtq.ent.gz | 90.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6vtq.json.gz 6vtq.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/vt/6vtq https://data.pdbj.org/pub/pdb/validation_reports/vt/6vtq ftp://data.pdbj.org/pub/pdb/validation_reports/vt/6vtq ftp://data.pdbj.org/pub/pdb/validation_reports/vt/6vtq | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6vtoC 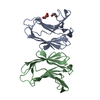 6vtpC  6vtrC  6vtsC  4galS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 15027.941 Da / Num. of mol.: 2 / Mutation: G16C Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: LGALS7, PIG1, LGALS7B / Production host: Homo sapiens (human) / Gene: LGALS7, PIG1, LGALS7B / Production host:  #2: Polysaccharide | #3: Chemical | #4: Water | ChemComp-HOH / | Has ligand of interest | Y | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.87 Å3/Da / Density % sol: 34.15 % |
|---|---|
| Crystal grow | Temperature: 295.15 K / Method: vapor diffusion, hanging drop / pH: 8 Details: 0.1 M Tris pH 8, 0.1 M Sodium Chloride, 25 % PEG3350, 7.5 mM alpha-lactose Temp details: Room temperature |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  CLSI CLSI  / Beamline: 08ID-1 / Wavelength: 0.97949 Å / Beamline: 08ID-1 / Wavelength: 0.97949 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: DECTRIS PILATUS3 S 6M / Detector: PIXEL / Date: Sep 28, 2019 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Monochromator: ACCEL/BRUKER double crystal mocochromator (DCM,Si-111) Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 0.97949 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.95→42.66 Å / Num. obs: 17138 / % possible obs: 100 % / Redundancy: 12.59 % / CC1/2: 0.999 / Rmerge(I) obs: 0.107 / Rrim(I) all: 0.112 / Net I/σ(I): 16.7 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
-Phasing
| Phasing | Method:  molecular replacement molecular replacement | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phasing MR |
|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 4gal Resolution: 1.95→42.66 Å / SU ML: 0.26 / Cross valid method: THROUGHOUT / σ(F): 1.35 / Phase error: 25.06 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 132.73 Å2 / Biso mean: 36.6079 Å2 / Biso min: 11.87 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 1.95→42.66 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Rfactor Rfree error: 0 / Total num. of bins used: 10
|
 Movie
Movie Controller
Controller












 PDBj
PDBj





