+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6ugg | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of unmodified E. coli tRNA(Asp) | |||||||||||||||
 Components Components | tRNAasp | |||||||||||||||
 Keywords Keywords | RNA / tRNA RNA unmodified | |||||||||||||||
| Function / homology | : / RNA / RNA (> 10) Function and homology information Function and homology information | |||||||||||||||
| Biological species |  | |||||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.95 Å MOLECULAR REPLACEMENT / Resolution: 1.95 Å | |||||||||||||||
 Authors Authors | Chan, C.W. / Mondragon, A. | |||||||||||||||
| Funding support |  United States, 4items United States, 4items
| |||||||||||||||
 Citation Citation |  Journal: Rna / Year: 2020 Journal: Rna / Year: 2020Title: Crystal structures of an unmodified bacterial tRNA reveal intrinsic structural flexibility and plasticity as general properties of unbound tRNAs. Authors: Chan, C.W. / Badong, D. / Rajan, R. / Mondragon, A. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6ugg.cif.gz 6ugg.cif.gz | 92.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6ugg.ent.gz pdb6ugg.ent.gz | 69.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6ugg.json.gz 6ugg.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  6ugg_validation.pdf.gz 6ugg_validation.pdf.gz | 247.9 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  6ugg_full_validation.pdf.gz 6ugg_full_validation.pdf.gz | 247.9 KB | Display | |
| Data in XML |  6ugg_validation.xml.gz 6ugg_validation.xml.gz | 940 B | Display | |
| Data in CIF |  6ugg_validation.cif.gz 6ugg_validation.cif.gz | 2.3 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ug/6ugg https://data.pdbj.org/pub/pdb/validation_reports/ug/6ugg ftp://data.pdbj.org/pub/pdb/validation_reports/ug/6ugg ftp://data.pdbj.org/pub/pdb/validation_reports/ug/6ugg | HTTPS FTP |
-Related structure data
| Related structure data | 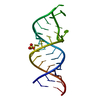 6ugiC  6ugjC  1ehzS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: RNA chain | Mass: 24815.684 Da / Num. of mol.: 2 / Source method: obtained synthetically / Source: (synth.)  #2: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.38 Å3/Da / Density % sol: 48.36 % Description: Clusters of arrow-like crystals. Single crystals excised from cluster. |
|---|---|
| Crystal grow | Temperature: 287 K / Method: vapor diffusion, hanging drop / pH: 7.5 Details: 2 uL 20 mg/ml E. coli tRNA(Asp) + 2 uL equilibration solution (50 mM sodium cacodylate, pH 7.5, 30% MPD, 10 mM spermine tetrahydrochloride, 300 mM sodium chloride, 300 mM potassium chloride) ...Details: 2 uL 20 mg/ml E. coli tRNA(Asp) + 2 uL equilibration solution (50 mM sodium cacodylate, pH 7.5, 30% MPD, 10 mM spermine tetrahydrochloride, 300 mM sodium chloride, 300 mM potassium chloride) + 0.5 uL 0.25% w/v p-coumaric acid, 0.25% w/v phenylurea, 0.25% w/v poly(3-hydroxybutyric acid), 0.25% w/v sulfaguanidine, 0.02 M sodium HEPES, pH 6.8 |
-Data collection
| Diffraction | Mean temperature: 100 K Ambient temp details: Crystals were flash frozen directly in mother liquor with liquid nitrogen Serial crystal experiment: N | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 21-ID-D / Wavelength: 1.0782 Å / Beamline: 21-ID-D / Wavelength: 1.0782 Å | |||||||||||||||
| Detector | Type: DECTRIS EIGER X 9M / Detector: PIXEL / Date: Dec 14, 2017 / Details: Monochromator | |||||||||||||||
| Radiation | Monochromator: Si(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||
| Radiation wavelength | Wavelength: 1.0782 Å / Relative weight: 1 | |||||||||||||||
| Reflection twin |
| |||||||||||||||
| Reflection | Resolution: 1.95→48.62 Å / Num. obs: 35638 / % possible obs: 94 % / Redundancy: 30 % / CC1/2: 0.992 / Rmerge(I) obs: 0.155 / Rpim(I) all: 0.029 / Rrim(I) all: 0.158 / Net I/σ(I): 15.8 | |||||||||||||||
| Reflection shell | Resolution: 1.95→2.056 Å / Redundancy: 33.7 % / Rmerge(I) obs: 2.683 / Mean I/σ(I) obs: 1.8 / Num. unique obs: 55102 / CC1/2: 0.488 / Rpim(I) all: 0.468 / Rrim(I) all: 2.724 / % possible all: 50.8 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB entry 1EHZ Resolution: 1.95→48.62 Å / Cor.coef. Fo:Fc: 0.941 / Cor.coef. Fo:Fc free: 0.929 / SU B: 2.442 / SU ML: 0.075 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.042 / ESU R Free: 0.035 Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS U VALUES : REFINED INDIVIDUALLY
| ||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å | ||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 123.64 Å2 / Biso mean: 50.999 Å2 / Biso min: 22.58 Å2
| ||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 1.95→48.62 Å
| ||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.95→2.001 Å / Rfactor Rfree error: 0 / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller













 PDBj
PDBj






























