[English] 日本語
 Yorodumi
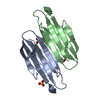
Yorodumi- PDB-6ucd: The crystal structure of Staphylococcus aureus super antigen-like... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6ucd | ||||||
|---|---|---|---|---|---|---|---|
| Title | The crystal structure of Staphylococcus aureus super antigen-like protein SSL10 | ||||||
 Components Components | Exotoxin | ||||||
 Keywords Keywords | TOXIN / Complement coagulation / IgG / Staphylococcus aureus / immune evasion | ||||||
| Function / homology |  Function and homology information Function and homology information | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 2.85 Å molecular replacement / Resolution: 2.85 Å | ||||||
 Authors Authors | Patel, D. / Young, P.G. / Bunker, R.D. / Baker, E.N. / Fraser, J.D. | ||||||
| Funding support |  New Zealand, 1items New Zealand, 1items
| ||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: The crystal structure of Staphylococcus aureus super antigen-like protein SSL10 Authors: Patel, D. / Hou, W. / Langley, R.J. / Young, P.G. / Ivanovic, I. / Bunker, R.D. / Baker, E.N. / Fraser, J.D. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6ucd.cif.gz 6ucd.cif.gz | 80.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6ucd.ent.gz pdb6ucd.ent.gz | 59.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6ucd.json.gz 6ucd.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/uc/6ucd https://data.pdbj.org/pub/pdb/validation_reports/uc/6ucd ftp://data.pdbj.org/pub/pdb/validation_reports/uc/6ucd ftp://data.pdbj.org/pub/pdb/validation_reports/uc/6ucd | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1v1oS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 23265.064 Da / Num. of mol.: 2 / Fragment: UNP residues 31-227 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Gene: tst, set14_2, BTN44_10650, EP54_13120, ER624_09120, HMPREF3211_02406, M1K003_2331, NCTC10654_00508, NCTC13131_05891, RK64_02640 Plasmid: pET32a / Production host:  |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.2 Å3/Da / Density % sol: 44.1 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, sitting drop / pH: 8.5 / Details: 30% PEG1500, 8% MPD, 0.1 M Tris, pH 8.5 |
-Data collection
| Diffraction | Mean temperature: 110 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU MICROMAX-007 HF / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU MICROMAX-007 HF / Wavelength: 1.5418 Å |
| Detector | Type: MAR scanner 345 mm plate / Detector: IMAGE PLATE / Date: Feb 16, 2010 |
| Radiation | Monochromator: osmic mirrors / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2.85→64.74 Å / Num. obs: 8204 / % possible obs: 89.5 % / Redundancy: 2.8 % / CC1/2: 0.98 / Rmerge(I) obs: 0.121 / Rpim(I) all: 0.081 / Rrim(I) all: 0.147 / Net I/av σ(I): 6.8 / Net I/σ(I): 6.8 |
| Reflection shell | Resolution: 2.85→3 Å / Redundancy: 2.5 % / Rmerge(I) obs: 0.557 / Mean I/σ(I) obs: 1.6 / Num. unique obs: 1139 / CC1/2: 0.44 / Rpim(I) all: 0.385 / Rrim(I) all: 0.683 / % possible all: 87.4 |
-Phasing
| Phasing | Method:  molecular replacement molecular replacement |
|---|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB entry 1V1O Resolution: 2.85→45.78 Å / Cross valid method: THROUGHOUT Details: TWIN DETAILS NUMBER OF TWIN DOMAINS : 2 TWIN DOMAIN : 1 TWIN OPERATOR : H, K, L TWIN FRACTION : 0.486 TWIN DOMAIN : 2 TWIN OPERATOR : -K, -H, -L TWIN FRACTION : 0.514
| ||||||||||||||||||||||||
| Displacement parameters | Biso max: 78.95 Å2 / Biso mean: 37.304 Å2 / Biso min: 16.41 Å2 | ||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.85→45.78 Å
| ||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.85→2.92 Å
|
 Movie
Movie Controller
Controller











 PDBj
PDBj
