+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6rp6 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Fragment AZ-019 binding at the TAZpS89/14-3-3 sigma interface | ||||||
 Components Components |
| ||||||
 Keywords Keywords | PEPTIDE BINDING PROTEIN / protein protein interaction / fragment soaking / stabilization | ||||||
| Function / homology |  Function and homology information Function and homology informationkidney morphogenesis / regulation of metanephric nephron tubule epithelial cell differentiation / RUNX3 regulates YAP1-mediated transcription / Physiological factors / mesenchymal cell differentiation / YAP1- and WWTR1 (TAZ)-stimulated gene expression / heart process / hippo signaling / tissue homeostasis / SMAD protein signal transduction ...kidney morphogenesis / regulation of metanephric nephron tubule epithelial cell differentiation / RUNX3 regulates YAP1-mediated transcription / Physiological factors / mesenchymal cell differentiation / YAP1- and WWTR1 (TAZ)-stimulated gene expression / heart process / hippo signaling / tissue homeostasis / SMAD protein signal transduction / EGR2 and SOX10-mediated initiation of Schwann cell myelination / glomerulus development / Signaling by Hippo / stem cell division / SCF-dependent proteasomal ubiquitin-dependent protein catabolic process / regulation of epidermal cell division / protein kinase C inhibitor activity / negative regulation of fat cell differentiation / positive regulation of epidermal cell differentiation / keratinocyte development / keratinization / RUNX2 regulates osteoblast differentiation / regulation of cell-cell adhesion / establishment of skin barrier / Regulation of localization of FOXO transcription factors / keratinocyte proliferation / cilium assembly / Activation of BAD and translocation to mitochondria / phosphoserine residue binding / negative regulation of keratinocyte proliferation / bicellular tight junction / cAMP/PKA signal transduction / negative regulation of protein localization to plasma membrane / positive regulation of osteoblast differentiation / positive regulation of epithelial to mesenchymal transition / SARS-CoV-2 targets host intracellular signalling and regulatory pathways / negative regulation of protein kinase activity / negative regulation of stem cell proliferation / SARS-CoV-1 targets host intracellular signalling and regulatory pathways / RHO GTPases activate PKNs / Chk1/Chk2(Cds1) mediated inactivation of Cyclin B:Cdk1 complex / positive regulation of protein localization / positive regulation of cell adhesion / protein sequestering activity / negative regulation of innate immune response / TP53 Regulates Transcription of Genes Involved in G2 Cell Cycle Arrest / protein export from nucleus / release of cytochrome c from mitochondria / positive regulation of protein export from nucleus / stem cell proliferation / TP53 Regulates Metabolic Genes / transcription coregulator activity / Translocation of SLC2A4 (GLUT4) to the plasma membrane / Downregulation of SMAD2/3:SMAD4 transcriptional activity / SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription / negative regulation of canonical Wnt signaling pathway / positive regulation of protein localization to nucleus / multicellular organism growth / intrinsic apoptotic signaling pathway in response to DNA damage / osteoblast differentiation / intracellular protein localization / transcription corepressor activity / sperm midpiece / regulation of protein localization / positive regulation of cell growth / transcription regulator complex / transcription coactivator activity / regulation of cell cycle / nuclear body / protein ubiquitination / cadherin binding / positive regulation of cell population proliferation / regulation of DNA-templated transcription / protein kinase binding / negative regulation of transcription by RNA polymerase II / signal transduction / protein homodimerization activity / positive regulation of transcription by RNA polymerase II / extracellular space / extracellular exosome / nucleoplasm / identical protein binding / nucleus / plasma membrane / cytoplasm / cytosol Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 1.885 Å MOLECULAR REPLACEMENT / Resolution: 1.885 Å | ||||||
 Authors Authors | Genet, S. / Wolter, M. / Guillory, X. / Somsen, B. / Leysen, S. / Patel, J. / Castaldi, P. / Ottmann, C. | ||||||
| Funding support |  Netherlands, 1items Netherlands, 1items
| ||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2020 Journal: J.Med.Chem. / Year: 2020Title: Fragment-based Differential Targeting of PPI Stabilizer Interfaces. Authors: Guillory, X. / Wolter, M. / Leysen, S. / Neves, J.F. / Kuusk, A. / Genet, S. / Somsen, B. / Morrow, J.K. / Rivers, E. / van Beek, L. / Patel, J. / Goodnow, R. / Schoenherr, H. / Fuller, N. / ...Authors: Guillory, X. / Wolter, M. / Leysen, S. / Neves, J.F. / Kuusk, A. / Genet, S. / Somsen, B. / Morrow, J.K. / Rivers, E. / van Beek, L. / Patel, J. / Goodnow, R. / Schoenherr, H. / Fuller, N. / Cao, Q. / Doveston, R.G. / Brunsveld, L. / Arkin, M.R. / Castaldi, P. / Boyd, H. / Landrieu, I. / Chen, H. / Ottmann, C. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6rp6.cif.gz 6rp6.cif.gz | 115.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6rp6.ent.gz pdb6rp6.ent.gz | 87.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6rp6.json.gz 6rp6.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/rp/6rp6 https://data.pdbj.org/pub/pdb/validation_reports/rp/6rp6 ftp://data.pdbj.org/pub/pdb/validation_reports/rp/6rp6 ftp://data.pdbj.org/pub/pdb/validation_reports/rp/6rp6 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6r5lC  6rhcC  6rjlC  6rjqC  6rjzC  6rk8C  6rkiC  6rkkC  6rkmC  6rl3C  6rl4C  6rl6C  6rm5C  6rm7C  6rwhC  6rwiC  6rwsC  6rwuC  6rx2C  6s39C  6s3cC  6s40C  6s9qC  6sinC  6sioC  6sipC  6siqC  6slvC  6slwC  6slxC  3mhrS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein / Protein/peptide , 2 types, 2 molecules AP
| #1: Protein | Mass: 26542.914 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: SFN, HME1 / Production host: Homo sapiens (human) / Gene: SFN, HME1 / Production host:  |
|---|---|
| #2: Protein/peptide | Mass: 1422.439 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.)  Homo sapiens (human) / References: UniProt: Q9GZV5 Homo sapiens (human) / References: UniProt: Q9GZV5 |
-Non-polymers , 4 types, 339 molecules 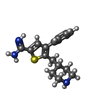






| #3: Chemical | ChemComp-KDK / |
|---|---|
| #4: Chemical | ChemComp-CA / |
| #5: Chemical | ChemComp-NA / |
| #6: Water | ChemComp-HOH / |
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.43 Å3/Da / Density % sol: 49.4 % |
|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop / pH: 7.1 Details: 0.095 M Na-HEPES pH 7.1, 27% PEG400, 0.19 M Calcium chloride, 5% Glycerol |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source: SEALED TUBE / Type: RIGAKU MICROMAX-003 / Wavelength: 1.54187 Å |
| Detector | Type: DECTRIS PILATUS 200K / Detector: PIXEL / Date: Aug 2, 2017 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.54187 Å / Relative weight: 1 |
| Reflection | Resolution: 1.62→41.86 Å / Num. obs: 31371 / % possible obs: 83.4 % / Redundancy: 5.6 % / CC1/2: 0.998 / Rrim(I) all: 0.082 / Net I/σ(I): 14 |
| Reflection shell | Resolution: 1.62→1.64 Å / Redundancy: 1.2 % / Mean I/σ(I) obs: 1.1 / Num. unique obs: 111 / CC1/2: 0.862 / Rrim(I) all: 0.391 / % possible all: 6.1 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3MHR Resolution: 1.885→25.576 Å / SU ML: 0.19 / Cross valid method: FREE R-VALUE / σ(F): 1.35 / Phase error: 20.73
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.885→25.576 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller





















 PDBj
PDBj





















