[English] 日本語
 Yorodumi
Yorodumi- PDB-6r4t: Crystal structure of the Pri1 subunit of human primase bound to v... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6r4t | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the Pri1 subunit of human primase bound to vidarabine triphosphate | ||||||
 Components Components | DNA primase small subunit | ||||||
 Keywords Keywords | REPLICATION / Primase / DNA-dependent RNA polymerase / ATP / priming | ||||||
| Function / homology |  Function and homology information Function and homology informationDNA primase AEP / ribonucleotide binding / DNA replication initiation / Inhibition of replication initiation of damaged DNA by RB1/E2F1 / Telomere C-strand synthesis initiation / alpha DNA polymerase:primase complex / Polymerase switching / Processive synthesis on the lagging strand / Removal of the Flap Intermediate / DNA replication, synthesis of primer ...DNA primase AEP / ribonucleotide binding / DNA replication initiation / Inhibition of replication initiation of damaged DNA by RB1/E2F1 / Telomere C-strand synthesis initiation / alpha DNA polymerase:primase complex / Polymerase switching / Processive synthesis on the lagging strand / Removal of the Flap Intermediate / DNA replication, synthesis of primer / Polymerase switching on the C-strand of the telomere / Activation of the pre-replicative complex / DNA replication initiation / Defective pyroptosis / DNA-directed RNA polymerase activity / magnesium ion binding / zinc ion binding / nucleoplasm / membrane Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.35 Å MOLECULAR REPLACEMENT / Resolution: 2.35 Å | ||||||
 Authors Authors | Kilkenny, M.L. / Pellegrini, L. | ||||||
| Funding support |  United Kingdom, 1items United Kingdom, 1items
| ||||||
 Citation Citation |  Journal: Acs Chem.Biol. / Year: 2019 Journal: Acs Chem.Biol. / Year: 2019Title: Structural Basis for Inhibition of Human Primase by Arabinofuranosyl Nucleoside Analogues Fludarabine and Vidarabine. Authors: Holzer, S. / Rzechorzek, N.J. / Short, I.R. / Jenkyn-Bedford, M. / Pellegrini, L. / Kilkenny, M.L. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6r4t.cif.gz 6r4t.cif.gz | 493.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6r4t.ent.gz pdb6r4t.ent.gz | 400.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6r4t.json.gz 6r4t.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/r4/6r4t https://data.pdbj.org/pub/pdb/validation_reports/r4/6r4t ftp://data.pdbj.org/pub/pdb/validation_reports/r4/6r4t ftp://data.pdbj.org/pub/pdb/validation_reports/r4/6r4t | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6r4sC  6r4uC  6r5dC  6r5eC  6rb4SC C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||
| 2 | 
| ||||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 2 molecules AD
| #1: Protein | Mass: 48720.516 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: PRIM1 / Production host: Homo sapiens (human) / Gene: PRIM1 / Production host:  References: UniProt: P49642, Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases |
|---|
-Non-polymers , 5 types, 71 molecules 

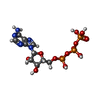






| #2: Chemical | | #3: Chemical | #4: Chemical | #5: Chemical | #6: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.56 Å3/Da / Density % sol: 51.87 % |
|---|---|
| Crystal grow | Temperature: 292 K / Method: vapor diffusion, sitting drop Details: 23% PEG3350, 10% ethylene glycol, 200 mM Na/K tartrate |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I24 / Wavelength: 0.97867 Å / Beamline: I24 / Wavelength: 0.97867 Å |
| Detector | Type: DECTRIS PILATUS3 6M / Detector: PIXEL / Date: May 3, 2015 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97867 Å / Relative weight: 1 |
| Reflection | Resolution: 2.35→46.691 Å / Num. obs: 41814 / % possible obs: 99.7 % / Redundancy: 7.2 % / CC1/2: 0.998 / Net I/σ(I): 15.35 |
| Reflection shell | Resolution: 2.35→2.49 Å / Mean I/σ(I) obs: 1.63 / Num. unique obs: 6609 / CC1/2: 0.81 / % possible all: 98.7 |
- Processing
Processing
| Software |
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 6RB4 Resolution: 2.35→46.69 Å / Cross valid method: FREE R-VALUE
| ||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.35→46.69 Å
|
 Movie
Movie Controller
Controller












 PDBj
PDBj








