[English] 日本語
 Yorodumi
Yorodumi- PDB-6puz: Structure of HIV cleaved synaptic complex (CSC) intasome bound wi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6puz | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of HIV cleaved synaptic complex (CSC) intasome bound with magnesium and INSTI XZ446 (compound 4f) | ||||||||||||
 Components Components |
| ||||||||||||
 Keywords Keywords | VIRAL PROTEIN/DNA / integrase / intasome / transposition / VIRAL PROTEIN-DNA complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationRNA endonuclease activity / DNA integration / viral genome integration into host DNA / establishment of integrated proviral latency / RNA stem-loop binding / viral penetration into host nucleus / RNA-directed DNA polymerase activity / host cell / viral nucleocapsid / endonuclease activity ...RNA endonuclease activity / DNA integration / viral genome integration into host DNA / establishment of integrated proviral latency / RNA stem-loop binding / viral penetration into host nucleus / RNA-directed DNA polymerase activity / host cell / viral nucleocapsid / endonuclease activity / DNA recombination / aspartic-type endopeptidase activity / DNA-directed DNA polymerase activity / host cell cytoplasm / symbiont-mediated suppression of host gene expression / viral translational frameshifting / symbiont entry into host cell / lipid binding / host cell nucleus / host cell plasma membrane / proteolysis / DNA binding / zinc ion binding / membrane / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |   Saccharolobus solfataricus (archaea) Saccharolobus solfataricus (archaea)  Human immunodeficiency virus 1 Human immunodeficiency virus 1 | ||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.8 Å | ||||||||||||
 Authors Authors | Lyumkis, D. / Jozwik, I.K. / Passos, D. | ||||||||||||
| Funding support |  United States, 3items United States, 3items
| ||||||||||||
 Citation Citation |  Journal: Science / Year: 2020 Journal: Science / Year: 2020Title: Structural basis for strand-transfer inhibitor binding to HIV intasomes. Authors: Dario Oliveira Passos / Min Li / Ilona K Jóźwik / Xue Zhi Zhao / Diogo Santos-Martins / Renbin Yang / Steven J Smith / Youngmin Jeon / Stefano Forli / Stephen H Hughes / Terrence R Burke / ...Authors: Dario Oliveira Passos / Min Li / Ilona K Jóźwik / Xue Zhi Zhao / Diogo Santos-Martins / Renbin Yang / Steven J Smith / Youngmin Jeon / Stefano Forli / Stephen H Hughes / Terrence R Burke / Robert Craigie / Dmitry Lyumkis /  Abstract: The HIV intasome is a large nucleoprotein assembly that mediates the integration of a DNA copy of the viral genome into host chromatin. Intasomes are targeted by the latest generation of ...The HIV intasome is a large nucleoprotein assembly that mediates the integration of a DNA copy of the viral genome into host chromatin. Intasomes are targeted by the latest generation of antiretroviral drugs, integrase strand-transfer inhibitors (INSTIs). Challenges associated with lentiviral intasome biochemistry have hindered high-resolution structural studies of how INSTIs bind to their native drug target. Here, we present high-resolution cryo-electron microscopy structures of HIV intasomes bound to the latest generation of INSTIs. These structures highlight how small changes in the integrase active site can have notable implications for drug binding and design and provide mechanistic insights into why a leading INSTI retains efficacy against a broad spectrum of drug-resistant variants. The data have implications for expanding effective treatments available for HIV-infected individuals. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6puz.cif.gz 6puz.cif.gz | 177.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6puz.ent.gz pdb6puz.ent.gz | 126.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6puz.json.gz 6puz.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/pu/6puz https://data.pdbj.org/pub/pdb/validation_reports/pu/6puz ftp://data.pdbj.org/pub/pdb/validation_reports/pu/6puz ftp://data.pdbj.org/pub/pdb/validation_reports/pu/6puz | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  20485MC  6putC  6puwC  6puyC  6v3kC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 | 
|
| 2 |
|
| 3 | 
|
| Symmetry | Point symmetry: (Schoenflies symbol: C2 (2 fold cyclic)) |
- Components
Components
-Protein , 1 types, 4 molecules ABCD
| #1: Protein | Mass: 42321.258 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2) (archaea), (gene. exp.) Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2) (archaea), (gene. exp.)   Human immunodeficiency virus 1 Human immunodeficiency virus 1Strain: ATCC 35092 / DSM 1617 / JCM 11322 / P2 / Gene: sso7d, sso7d-1, SSO10610 / Production host:  |
|---|
-DNA chain , 2 types, 2 molecules EF
| #2: DNA chain | Mass: 8188.271 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.)   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
|---|---|
| #3: DNA chain | Mass: 7773.023 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.)   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Non-polymers , 4 types, 175 molecules 

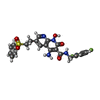




| #4: Chemical | | #5: Chemical | #6: Chemical | ChemComp-XXJ / | #7: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Assembly of HIV integrase and viral DNA / Type: COMPLEX / Entity ID: #1-#3 / Source: RECOMBINANT | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Value: 0.4 MDa / Experimental value: NO | ||||||||||||||||||||||||||||||
| Source (natural) | Organism:   hiv (virus) hiv (virus) | ||||||||||||||||||||||||||||||
| Source (recombinant) | Organism:  | ||||||||||||||||||||||||||||||
| Buffer solution | pH: 6.2 | ||||||||||||||||||||||||||||||
| Buffer component |
| ||||||||||||||||||||||||||||||
| Specimen | Conc.: 0.2 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | ||||||||||||||||||||||||||||||
| Specimen support | Grid material: GOLD / Grid mesh size: 400 divisions/in. / Grid type: UltrAuFoil | ||||||||||||||||||||||||||||||
| Vitrification | Instrument: HOMEMADE PLUNGER / Cryogen name: ETHANE / Humidity: 80 % |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 37000 X / Calibrated magnification: 63291 X / Nominal defocus max: 3000 nm / Nominal defocus min: 1500 nm / Cs: 2.7 mm / C2 aperture diameter: 70 µm / Alignment procedure: COMA FREE |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Electron dose: 42 e/Å2 / Detector mode: COUNTING / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Num. of real images: 2072 |
| Image scans | Width: 3838 / Height: 3710 / Movie frames/image: 80 / Used frames/image: 4-80 |
- Processing
Processing
| Software | Name: PHENIX / Version: (1.16_3549: ???) / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 412354 | ||||||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: C2 (2 fold cyclic) | ||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 2.8 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 143698 / Algorithm: FOURIER SPACE / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: FLEXIBLE FIT / Space: REAL / Target criteria: Correlation coefficient | ||||||||||||||||||||||||||||||||||||||||
| Atomic model building |
| ||||||||||||||||||||||||||||||||||||||||
| Refinement | Resolution: 2.8→303.36 Å / SU ML: 1.19 / Phase error: 61.61 / Stereochemistry target values: ML
| ||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj












































