+Search query
-Structure paper
| Title | Structural basis for strand-transfer inhibitor binding to HIV intasomes. |
|---|---|
| Journal, issue, pages | Science, Vol. 367, Issue 6479, Page 810-814, Year 2020 |
| Publish date | Feb 14, 2020 |
 Authors Authors | Dario Oliveira Passos / Min Li / Ilona K Jóźwik / Xue Zhi Zhao / Diogo Santos-Martins / Renbin Yang / Steven J Smith / Youngmin Jeon / Stefano Forli / Stephen H Hughes / Terrence R Burke / Robert Craigie / Dmitry Lyumkis /  |
| PubMed Abstract | The HIV intasome is a large nucleoprotein assembly that mediates the integration of a DNA copy of the viral genome into host chromatin. Intasomes are targeted by the latest generation of ...The HIV intasome is a large nucleoprotein assembly that mediates the integration of a DNA copy of the viral genome into host chromatin. Intasomes are targeted by the latest generation of antiretroviral drugs, integrase strand-transfer inhibitors (INSTIs). Challenges associated with lentiviral intasome biochemistry have hindered high-resolution structural studies of how INSTIs bind to their native drug target. Here, we present high-resolution cryo-electron microscopy structures of HIV intasomes bound to the latest generation of INSTIs. These structures highlight how small changes in the integrase active site can have notable implications for drug binding and design and provide mechanistic insights into why a leading INSTI retains efficacy against a broad spectrum of drug-resistant variants. The data have implications for expanding effective treatments available for HIV-infected individuals. |
 External links External links |  Science / Science /  PubMed:32001521 / PubMed:32001521 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.8 - 3.4 Å |
| Structure data | EMDB-20481, PDB-6put: EMDB-20483, PDB-6puw: EMDB-20484, PDB-6puy: EMDB-20485, PDB-6puz: EMDB-21038, PDB-6v3k: |
| Chemicals |  ChemComp-CA:  ChemComp-ZN:  ChemComp-HOH:  ChemComp-MG: 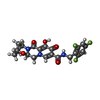 ChemComp-KLQ: 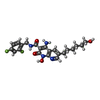 ChemComp-OZ1: 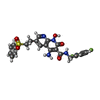 ChemComp-XXJ: 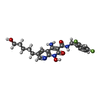 ChemComp-QUW: |
| Source |
|
 Keywords Keywords | VIRAL PROTEIN/DNA / integrase / intasome / enzyme / transposition / VIRAL PROTEIN / VIRAL PROTEIN-DNA complex |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers














 human immunodeficiency virus 1
human immunodeficiency virus 1
 saccharolobus solfataricus (strain atcc 35092 / dsm 1617 / jcm 11322 / p2) (archaea)
saccharolobus solfataricus (strain atcc 35092 / dsm 1617 / jcm 11322 / p2) (archaea)