| Software | | Name | Version | Classification |
|---|
| PHENIX | 1.13 | refinement| PDB_EXTRACT | 3.24 | data extraction| XDS | | data reduction| XSCALE | | data scaling| Aimless | | data scaling| HKL2Map | | phasing | | | | | |
|
|---|
| Refinement | Method to determine structure:  SAD / Resolution: 1.47→23.8 Å / SU ML: 0.14 / Cross valid method: THROUGHOUT / σ(F): 1.33 / Phase error: 29.86 / Stereochemistry target values: ML SAD / Resolution: 1.47→23.8 Å / SU ML: 0.14 / Cross valid method: THROUGHOUT / σ(F): 1.33 / Phase error: 29.86 / Stereochemistry target values: ML
| Rfactor | Num. reflection | % reflection |
|---|
| Rfree | 0.1992 | 610 | 4.81 % |
|---|
| Rwork | 0.1921 | 12073 | - |
|---|
| obs | 0.1924 | 12683 | 98.04 % |
|---|
|
|---|
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL |
|---|
| Displacement parameters | Biso max: 91.76 Å2 / Biso mean: 33.7619 Å2 / Biso min: 19.85 Å2 |
|---|
| Refinement step | Cycle: final / Resolution: 1.47→23.8 Å
| Protein | Nucleic acid | Ligand | Solvent | Total |
|---|
| Num. atoms | 0 | 580 | 25 | 67 | 672 |
|---|
| Biso mean | - | - | 50.68 | 39.21 | - |
|---|
| Num. residues | - | - | - | - | 28 |
|---|
|
|---|
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Rfactor Rfree error: 0 | Resolution (Å) | Rfactor Rfree | Num. reflection Rfree | Rfactor Rwork | Num. reflection Rwork | % reflection obs (%) |
|---|
| 1.47-1.6179 | 0.3471 | 121 | 0.2829 | 2917 | 94 | | 1.6179-1.852 | 0.2591 | 156 | 0.2065 | 3023 | 99 | | 1.852-2.333 | 0.2218 | 179 | 0.2147 | 3043 | 100 | | 2.333-23.8 | 0.1736 | 154 | 0.1772 | 3090 | 99 |
|
|---|
| Refinement TLS params. | Method: refined / Origin x: -3.603 Å / Origin y: -7.6468 Å / Origin z: -7.6421 Å
| 11 | 12 | 13 | 21 | 22 | 23 | 31 | 32 | 33 |
|---|
| T | 0.2338 Å2 | -0.0152 Å2 | -0.0058 Å2 | - | 0.2214 Å2 | -0.0077 Å2 | - | - | 0.2398 Å2 |
|---|
| L | 1.7307 °2 | -0.6688 °2 | -0.6631 °2 | - | 1.4693 °2 | -0.101 °2 | - | - | 2.0666 °2 |
|---|
| S | -0.0404 Å ° | 0.1328 Å ° | -0.1068 Å ° | -0.0897 Å ° | 0.0395 Å ° | 0.0078 Å ° | -0.0505 Å ° | 0.048 Å ° | -0.0005 Å ° |
|---|
|
|---|
| Refinement TLS group | | ID | Refine-ID | Refine TLS-ID | Selection details | Auth asym-ID | Auth seq-ID |
|---|
| 1 | X-RAY DIFFRACTION | 1 | allA| 1 - 14 | | 2 | X-RAY DIFFRACTION | 1 | allB| 1 - 14 | | 3 | X-RAY DIFFRACTION | 1 | allC| 1 - 3 | | 4 | X-RAY DIFFRACTION | 1 | allD| 1 | | 5 | X-RAY DIFFRACTION | 1 | allE| 101 - 103 | | 6 | X-RAY DIFFRACTION | 1 | allS| 1 - 68 | | | | | | | | | | | | |
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
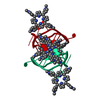
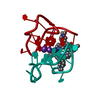
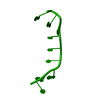
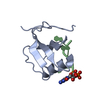
Function and homology information Pseudorabies virus Ea
Pseudorabies virus Ea X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  SAD / Resolution: 1.47 Å
SAD / Resolution: 1.47 Å  Authors
Authors China, 1items
China, 1items  Citation
Citation Journal: Nucleic Acids Res. / Year: 2020
Journal: Nucleic Acids Res. / Year: 2020 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6jjf.cif.gz
6jjf.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6jjf.ent.gz
pdb6jjf.ent.gz PDB format
PDB format 6jjf.json.gz
6jjf.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/jj/6jjf
https://data.pdbj.org/pub/pdb/validation_reports/jj/6jjf ftp://data.pdbj.org/pub/pdb/validation_reports/jj/6jjf
ftp://data.pdbj.org/pub/pdb/validation_reports/jj/6jjf Links
Links Assembly
Assembly
 Components
Components Pseudorabies virus Ea
Pseudorabies virus Ea X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation Processing
Processing SAD / Resolution: 1.47→23.8 Å / SU ML: 0.14 / Cross valid method: THROUGHOUT / σ(F): 1.33 / Phase error: 29.86 / Stereochemistry target values: ML
SAD / Resolution: 1.47→23.8 Å / SU ML: 0.14 / Cross valid method: THROUGHOUT / σ(F): 1.33 / Phase error: 29.86 / Stereochemistry target values: ML Movie
Movie Controller
Controller








 PDBj
PDBj















































