[English] 日本語
 Yorodumi
Yorodumi- PDB-6jjh: Crystal structure of a two-quartet RNA parallel G-quadruplex comp... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6jjh | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of a two-quartet RNA parallel G-quadruplex complexed with the porphyrin TMPyP4 | ||||||||||||||||||||||||||||
 Components Components | RNA (5'-R(* Keywords KeywordsRNA / G-quadruplex / two-quartet / complex | Function / homology | : / Chem-POH / RNA / RNA (> 10) |  Function and homology information Function and homology informationBiological species |  Pseudorabies virus Ea Pseudorabies virus EaMethod |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  SAD / Resolution: 1.74 Å SAD / Resolution: 1.74 Å  Authors AuthorsZhang, Y.S. / EI Omari, K. / Duman, R. / Wagner, A. / Parkinson, G.N. / Wei, D.G. | Funding support | |  China, 1items China, 1items
 Citation Citation Journal: Nucleic Acids Res. / Year: 2020 Journal: Nucleic Acids Res. / Year: 2020Title: Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths. Authors: Zhang, Y. / El Omari, K. / Duman, R. / Liu, S. / Haider, S. / Wagner, A. / Parkinson, G.N. / Wei, D. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6jjh.cif.gz 6jjh.cif.gz | 23.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6jjh.ent.gz pdb6jjh.ent.gz | 14.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6jjh.json.gz 6jjh.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/jj/6jjh https://data.pdbj.org/pub/pdb/validation_reports/jj/6jjh ftp://data.pdbj.org/pub/pdb/validation_reports/jj/6jjh ftp://data.pdbj.org/pub/pdb/validation_reports/jj/6jjh | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 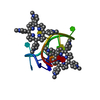
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 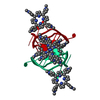
| ||||||||||||
| Unit cell |
| ||||||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: RNA chain | Mass: 4572.788 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.)  Pseudorabies virus Ea Pseudorabies virus Ea | ||||||
|---|---|---|---|---|---|---|---|
| #2: Chemical | | #3: Chemical | #4: Water | ChemComp-HOH / | Has ligand of interest | Y | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.29 Å3/Da / Density % sol: 46.17 % / Description: rod like crystals |
|---|---|
| Crystal grow | Temperature: 283 K / Method: vapor diffusion, hanging drop / pH: 6.5 Details: Potassium cacodylate, Potassium chloride, Sodium cacodylate, MPD |
-Data collection
| Diffraction |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source |
| ||||||||||||||||||||||||
| Detector |
| ||||||||||||||||||||||||
| Radiation |
| ||||||||||||||||||||||||
| Radiation wavelength |
| ||||||||||||||||||||||||
| Reflection | Entry-ID: 6JJH
| ||||||||||||||||||||||||
| Reflection shell |
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  SAD / Resolution: 1.74→29.6 Å / Cor.coef. Fo:Fc: 0.961 / Cor.coef. Fo:Fc free: 0.911 / SU B: 4.661 / SU ML: 0.137 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.176 / ESU R Free: 0.165 / Stereochemistry target values: MAXIMUM LIKELIHOOD SAD / Resolution: 1.74→29.6 Å / Cor.coef. Fo:Fc: 0.961 / Cor.coef. Fo:Fc free: 0.911 / SU B: 4.661 / SU ML: 0.137 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.176 / ESU R Free: 0.165 / Stereochemistry target values: MAXIMUM LIKELIHOODDetails: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS U VALUES : REFINED INDIVIDUALLY SF FILE CONTAINS FRIEDEL PAIRS UNDER I/F_MINUS AND I/F_PLUS COLUMNS.
| ||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 105.63 Å2 / Biso mean: 42.481 Å2 / Biso min: 21.04 Å2
| ||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 1.74→29.6 Å
| ||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.74→1.785 Å / Rfactor Rfree error: 0 / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller











 PDBj
PDBj