[English] 日本語
 Yorodumi
Yorodumi- PDB-6gfi: Structure of Human Mesotrypsin in complex with APPI variant T11V/... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6gfi | ||||||
|---|---|---|---|---|---|---|---|
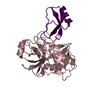
| Title | Structure of Human Mesotrypsin in complex with APPI variant T11V/M17R/I18F/F34V | ||||||
 Components Components |
| ||||||
 Keywords Keywords | PROTEIN BINDING / protein protein interactions / serine proteases / amyloid protein precursor inhibitor | ||||||
| Function / homology |  Function and homology information Function and homology informationUptake of dietary cobalamins into enterocytes / antimicrobial humoral response / Alpha-defensins / amyloid-beta complex / growth cone lamellipodium / cellular response to norepinephrine stimulus / growth cone filopodium / microglia development / collateral sprouting in absence of injury / Differentiation of Keratinocytes in Interfollicular Epidermis in Mammalian Skin ...Uptake of dietary cobalamins into enterocytes / antimicrobial humoral response / Alpha-defensins / amyloid-beta complex / growth cone lamellipodium / cellular response to norepinephrine stimulus / growth cone filopodium / microglia development / collateral sprouting in absence of injury / Differentiation of Keratinocytes in Interfollicular Epidermis in Mammalian Skin / Formyl peptide receptors bind formyl peptides and many other ligands / axo-dendritic transport / regulation of Wnt signaling pathway / regulation of synapse structure or activity / axon midline choice point recognition / zymogen activation / astrocyte activation involved in immune response / NMDA selective glutamate receptor signaling pathway / regulation of spontaneous synaptic transmission / mating behavior / Antimicrobial peptides / growth factor receptor binding / peptidase activator activity / Golgi-associated vesicle / PTB domain binding / positive regulation of amyloid fibril formation / Insertion of tail-anchored proteins into the endoplasmic reticulum membrane / astrocyte projection / Lysosome Vesicle Biogenesis / neuron remodeling / Deregulated CDK5 triggers multiple neurodegenerative pathways in Alzheimer's disease models / nuclear envelope lumen / dendrite development / positive regulation of protein metabolic process / TRAF6 mediated NF-kB activation / signaling receptor activator activity / negative regulation of long-term synaptic potentiation / Advanced glycosylation endproduct receptor signaling / modulation of excitatory postsynaptic potential / main axon / trypsin / transition metal ion binding / The NLRP3 inflammasome / regulation of multicellular organism growth / intracellular copper ion homeostasis / ECM proteoglycans / regulation of presynapse assembly / endothelial cell migration / positive regulation of T cell migration / neuronal dense core vesicle / Purinergic signaling in leishmaniasis infection / positive regulation of chemokine production / digestion / Notch signaling pathway / cellular response to manganese ion / clathrin-coated pit / extracellular matrix organization / neuron projection maintenance / astrocyte activation / serine-type peptidase activity / ionotropic glutamate receptor signaling pathway / Mitochondrial protein degradation / positive regulation of calcium-mediated signaling / positive regulation of mitotic cell cycle / axonogenesis / protein serine/threonine kinase binding / response to interleukin-1 / platelet alpha granule lumen / cellular response to copper ion / cellular response to cAMP / positive regulation of glycolytic process / central nervous system development / dendritic shaft / trans-Golgi network membrane / endosome lumen / positive regulation of long-term synaptic potentiation / adult locomotory behavior / positive regulation of interleukin-1 beta production / learning / Post-translational protein phosphorylation / positive regulation of JNK cascade / locomotory behavior / serine-type endopeptidase inhibitor activity / microglial cell activation / positive regulation of non-canonical NF-kappaB signal transduction / cellular response to nerve growth factor stimulus / TAK1-dependent IKK and NF-kappa-B activation / regulation of long-term neuronal synaptic plasticity / recycling endosome / synapse organization / visual learning / response to lead ion / positive regulation of interleukin-6 production / Golgi lumen / cognition / Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) / endocytosis / cellular response to amyloid-beta / neuron projection development / positive regulation of inflammatory response Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.3 Å MOLECULAR REPLACEMENT / Resolution: 2.3 Å | ||||||
 Authors Authors | Shahar, A. / Cohen, I. / Radisky, E. / Papo, N. / Naftaly, S. | ||||||
| Funding support |  Israel, 1items Israel, 1items
| ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2018 Journal: Nat Commun / Year: 2018Title: Mapping protein selectivity landscapes using multi-target selective screening and next-generation sequencing of combinatorial libraries. Authors: Naftaly, S. / Cohen, I. / Shahar, A. / Hockla, A. / Radisky, E.S. / Papo, N. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6gfi.cif.gz 6gfi.cif.gz | 233.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6gfi.ent.gz pdb6gfi.ent.gz | 189.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6gfi.json.gz 6gfi.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/gf/6gfi https://data.pdbj.org/pub/pdb/validation_reports/gf/6gfi ftp://data.pdbj.org/pub/pdb/validation_reports/gf/6gfi ftp://data.pdbj.org/pub/pdb/validation_reports/gf/6gfi | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5c67S S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 24257.457 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: PRSS3 Homo sapiens (human) / Gene: PRSS3Production host: References: UniProt: Q8N2U3, UniProt: P35030*PLUS #2: Protein | Mass: 9185.026 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: APP, A4, AD1 / Production host: Homo sapiens (human) / Gene: APP, A4, AD1 / Production host:  Komagataella phaffii GS115 (fungus) / References: UniProt: P05067 Komagataella phaffii GS115 (fungus) / References: UniProt: P05067#3: Chemical | ChemComp-EDO / | #4: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.9 Å3/Da / Density % sol: 57.57 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop Details: 0.1M Sodium Chloride, 0.1M Bis-Tris pH 6.8 , 1.42M Ammonium Sulfate |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID30B / Wavelength: 0.9762 Å / Beamline: ID30B / Wavelength: 0.9762 Å |
| Detector | Type: DECTRIS PILATUS3 S 6M / Detector: PIXEL / Date: Oct 27, 2017 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9762 Å / Relative weight: 1 |
| Reflection | Resolution: 2.3→46.09 Å / Num. obs: 33309 / % possible obs: 99.7 % / Redundancy: 8.3 % / Rpim(I) all: 0.033 / Net I/σ(I): 10.9 |
| Reflection shell | Resolution: 2.3→2.38 Å / Rpim(I) all: 0.269 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5C67 Resolution: 2.3→46.086 Å / SU ML: 0.42 / Cross valid method: FREE R-VALUE / σ(F): 1.96 / Phase error: 37.89
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.3→46.086 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj





















