Entry Database : PDB / ID : 5c67Title Human Mesotrypsin in complex with amyloid precursor protein inhibitor variant APPI-M17G/I18F/F34V Amyloid beta A4 protein Trypsin-3 Keywords / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / / / Resolution : 1.83 Å Authors Kayode, O. / Sankaran, B. / Radisky, E.S. Funding support Organization Grant number Country National Institutes of Health/National Cancer Institute (NIH/NCI) R01CA154387
Journal : Biochem.J. / Year : 2016Title : Combinatorial protein engineering of proteolytically resistant mesotrypsin inhibitors as candidates for cancer therapy.Authors : Cohen, I. / Kayode, O. / Hockla, A. / Sankaran, B. / Radisky, D.C. / Radisky, E.S. / Papo, N. History Deposition Jun 22, 2015 Deposition site / Processing site Revision 1.0 May 4, 2016 Provider / Type Revision 1.1 May 18, 2016 Group / Source and taxonomyRevision 1.2 Jun 1, 2016 Group Revision 1.3 Sep 6, 2017 Group / Database references / Derived calculationsCategory / pdbx_audit_support / pdbx_struct_oper_listItem / _pdbx_audit_support.funding_organization / _pdbx_struct_oper_list.symmetry_operationRevision 1.4 Dec 4, 2019 Group / Derived calculationsCategory / pdbx_struct_assembly_gen / pdbx_struct_assembly_propItem / _pdbx_struct_assembly_gen.asym_id_list / _pdbx_struct_assembly_prop.valueRevision 1.5 Sep 27, 2023 Group / Database references / Refinement descriptionCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model Item / _database_2.pdbx_database_accessionRevision 1.6 Oct 23, 2024 Group / Category / pdbx_modification_feature
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.83 Å
MOLECULAR REPLACEMENT / Resolution: 1.83 Å  Authors
Authors United States, 1items
United States, 1items  Citation
Citation Journal: Biochem.J. / Year: 2016
Journal: Biochem.J. / Year: 2016 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5c67.cif.gz
5c67.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5c67.ent.gz
pdb5c67.ent.gz PDB format
PDB format 5c67.json.gz
5c67.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/c6/5c67
https://data.pdbj.org/pub/pdb/validation_reports/c6/5c67 ftp://data.pdbj.org/pub/pdb/validation_reports/c6/5c67
ftp://data.pdbj.org/pub/pdb/validation_reports/c6/5c67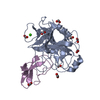
 Links
Links Assembly
Assembly


 Components
Components Homo sapiens (human) / Gene: PRSS3, PRSS4, TRY3, TRY4 / Organ: PANCREAS / Plasmid: pTRAP-T7-wtHU3 / Production host:
Homo sapiens (human) / Gene: PRSS3, PRSS4, TRY3, TRY4 / Organ: PANCREAS / Plasmid: pTRAP-T7-wtHU3 / Production host: 
 Homo sapiens (human) / Gene: APP, A4, AD1 / Plasmid: pPICZalpha-APPI / Production host:
Homo sapiens (human) / Gene: APP, A4, AD1 / Plasmid: pPICZalpha-APPI / Production host:  Komagataella pastoris GS115 (fungus) / Strain (production host): GS115 / References: UniProt: P05067
Komagataella pastoris GS115 (fungus) / Strain (production host): GS115 / References: UniProt: P05067 X-RAY DIFFRACTION
X-RAY DIFFRACTION Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  ALS
ALS  / Beamline: 8.2.1 / Wavelength: 1 Å
/ Beamline: 8.2.1 / Wavelength: 1 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller












 PDBj
PDBj



















