| Entry | Database: PDB / ID: 6g2r
|
|---|
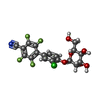
| Title | Crystal structure of FimH in complex with a tetraflourinated biphenyl alpha D-mannoside |
|---|
 Components Components | Type 1 fimbrin D-mannose specific adhesin |
|---|
 Keywords Keywords | CELL ADHESION / TYPE I PILUS / CATCH-BOND / LECTIN / UPEC / INFECTION / MANNOSE |
|---|
| Function / homology |  Function and homology information Function and homology information
pilus tip / mechanosensory behavior / cell adhesion involved in single-species biofilm formation / pilus / cell-substrate adhesion / D-mannose binding / host cell membrane / cell adhesionSimilarity search - Function Fimbrial-type adhesion domain / FimH, mannose-binding domain / FimH, mannose binding / Fimbrial-type adhesion domain / Fimbrial protein / : / Fimbrial-type adhesion domain superfamily / Adhesion domain superfamily / Immunoglobulin-like / Sandwich / Mainly BetaSimilarity search - Domain/homology |
|---|
| Biological species |   Escherichia coli (E. coli) Escherichia coli (E. coli) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.1 Å MOLECULAR REPLACEMENT / Resolution: 2.1 Å |
|---|
 Authors Authors | Jakob, R.P. / Schoenemann, W. / Cramer, J. / Muehlethaler, T. / Daetwyler, P. / Zihlmann, P. / Fiege, B. / Sager, C.P. / Smiesko, M. / Rabbani, S. ...Jakob, R.P. / Schoenemann, W. / Cramer, J. / Muehlethaler, T. / Daetwyler, P. / Zihlmann, P. / Fiege, B. / Sager, C.P. / Smiesko, M. / Rabbani, S. / Eris, D. / Schwardt, O. / Maier, T. / Ernst, B. |
|---|
 Citation Citation |  Journal: Chemmedchem / Year: 2019 Journal: Chemmedchem / Year: 2019
Title: Improvement of Aglycone pi-Stacking Yields Nanomolar to Sub-nanomolar FimH Antagonists.
Authors: Schonemann, W. / Cramer, J. / Muhlethaler, T. / Fiege, B. / Silbermann, M. / Rabbani, S. / Datwyler, P. / Zihlmann, P. / Jakob, R.P. / Sager, C.P. / Smiesko, M. / Schwardt, O. / Maier, T. / Ernst, B. |
|---|
| History | | Deposition | Mar 23, 2018 | Deposition site: PDBE / Processing site: PDBE |
|---|
| Revision 1.0 | Mar 20, 2019 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Apr 17, 2019 | Group: Data collection / Database references / Category: citation / citation_author / pdbx_database_proc
Item: _citation.journal_volume / _citation.page_first ..._citation.journal_volume / _citation.page_first / _citation.page_last / _citation_author.identifier_ORCID |
|---|
| Revision 1.2 | Jan 17, 2024 | Group: Data collection / Database references / Refinement description
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession |
|---|
| Revision 1.3 | Nov 6, 2024 | Group: Structure summary / Category: pdbx_entry_details / pdbx_modification_feature |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.1 Å
MOLECULAR REPLACEMENT / Resolution: 2.1 Å  Authors
Authors Citation
Citation Journal: Chemmedchem / Year: 2019
Journal: Chemmedchem / Year: 2019 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6g2r.cif.gz
6g2r.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6g2r.ent.gz
pdb6g2r.ent.gz PDB format
PDB format 6g2r.json.gz
6g2r.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/g2/6g2r
https://data.pdbj.org/pub/pdb/validation_reports/g2/6g2r ftp://data.pdbj.org/pub/pdb/validation_reports/g2/6g2r
ftp://data.pdbj.org/pub/pdb/validation_reports/g2/6g2r
 Links
Links Assembly
Assembly

 Components
Components

 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  SLS
SLS  / Beamline: X06SA / Wavelength: 1.87287 Å
/ Beamline: X06SA / Wavelength: 1.87287 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller




















 PDBj
PDBj