| Entry | Database: PDB / ID: 5cgb
|
|---|
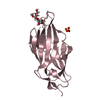
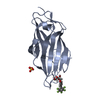
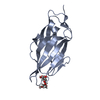
| Title | Crystal structure of FimH in complex with heptyl alpha-D-septanoside |
|---|
 Components Components | Protein FimH |
|---|
 Keywords Keywords | SUGAR BINDING PROTEIN / UTI / lectin / urinary tract infection / type 1 fimbriae / pilus / inhibitor |
|---|
| Function / homology |  Function and homology information Function and homology information
pilus tip / mechanosensory behavior / cell adhesion involved in single-species biofilm formation / pilus / cell-substrate adhesion / D-mannose binding / host cell membrane / cell adhesionSimilarity search - Function Fimbrial-type adhesion domain / FimH, mannose-binding domain / FimH, mannose binding / Fimbrial-type adhesion domain / Fimbrial protein / : / Fimbrial-type adhesion domain superfamily / Adhesion domain superfamily / Immunoglobulin-like / Sandwich / Mainly BetaSimilarity search - Domain/homology |
|---|
| Biological species |   Escherichia coli K-12 (bacteria) Escherichia coli K-12 (bacteria) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.6 Å MOLECULAR REPLACEMENT / Resolution: 1.6 Å |
|---|
 Authors Authors | Jakob, R.P. / Preston, R.P. / Zihlmann, P. / Fiege, B. / Sager, C.P. / Vannam, R. / Rabbani, S. / Zalewski, A. / Maier, T. / Ernst, B. / Peczuh, M. |
|---|
 Citation Citation |  Journal: Chem Sci / Year: 2018 Journal: Chem Sci / Year: 2018
Title: The price of flexibility - a case study on septanoses as pyranose mimetics.
Authors: Sager, C.P. / Fiege, B. / Zihlmann, P. / Vannam, R. / Rabbani, S. / Jakob, R.P. / Preston, R.C. / Zalewski, A. / Maier, T. / Peczuh, M.W. / Ernst, B. |
|---|
| History | | Deposition | Jul 9, 2015 | Deposition site: RCSB / Processing site: PDBE |
|---|
| Revision 1.0 | Jul 20, 2016 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Apr 7, 2021 | Group: Database references / Category: citation / citation_author
Item: _citation.country / _citation.journal_abbrev ..._citation.country / _citation.journal_abbrev / _citation.journal_id_CSD / _citation.journal_id_ISSN / _citation.journal_volume / _citation.page_first / _citation.page_last / _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation.year / _citation_author.identifier_ORCID / _citation_author.name |
|---|
| Revision 1.2 | Jan 10, 2024 | Group: Data collection / Database references / Refinement description
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession |
|---|
| Revision 1.3 | Nov 13, 2024 | Group: Structure summary / Category: pdbx_entry_details / pdbx_modification_feature |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.6 Å
MOLECULAR REPLACEMENT / Resolution: 1.6 Å  Authors
Authors Citation
Citation Journal: Chem Sci / Year: 2018
Journal: Chem Sci / Year: 2018 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5cgb.cif.gz
5cgb.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5cgb.ent.gz
pdb5cgb.ent.gz PDB format
PDB format 5cgb.json.gz
5cgb.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/cg/5cgb
https://data.pdbj.org/pub/pdb/validation_reports/cg/5cgb ftp://data.pdbj.org/pub/pdb/validation_reports/cg/5cgb
ftp://data.pdbj.org/pub/pdb/validation_reports/cg/5cgb
 Links
Links Assembly
Assembly


 Components
Components

 X-RAY DIFFRACTION
X-RAY DIFFRACTION Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  SLS
SLS  / Beamline: X06SA / Wavelength: 1 Å
/ Beamline: X06SA / Wavelength: 1 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller



















 PDBj
PDBj



