Entry Database : PDB / ID : 6a9cTitle Crystal Structure c-terminal SH3 domain of Myosin IB from Entamoeba histolytica bound to EhFP10(GEF) peptide. Peptide from Rho guanine nucleotide exchange factor Unconventional myosin IB Keywords / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Entamoeba histolytica (eukaryote)Method / / / Resolution : 1.98 Å Authors Gautam, G. / Gourinath, S. Journal : Plos Pathog. / Year : 2019Title : EhFP10: A FYVE family GEF interacts with myosin IB to regulate cytoskeletal dynamics during endocytosis in Entamoeba histolytica.Authors : Gautam, G. / Ali, M.S. / Bhattacharya, A. / Gourinath, S. History Deposition Jul 13, 2018 Deposition site / Processing site Revision 1.0 Jun 12, 2019 Provider / Type Revision 1.1 Nov 22, 2023 Group / Database references / Refinement descriptionCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model Item / _database_2.pdbx_database_accession
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.98 Å
MOLECULAR REPLACEMENT / Resolution: 1.98 Å  Authors
Authors Citation
Citation Journal: Plos Pathog. / Year: 2019
Journal: Plos Pathog. / Year: 2019 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6a9c.cif.gz
6a9c.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6a9c.ent.gz
pdb6a9c.ent.gz PDB format
PDB format 6a9c.json.gz
6a9c.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/a9/6a9c
https://data.pdbj.org/pub/pdb/validation_reports/a9/6a9c ftp://data.pdbj.org/pub/pdb/validation_reports/a9/6a9c
ftp://data.pdbj.org/pub/pdb/validation_reports/a9/6a9c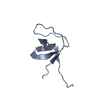
 Links
Links Assembly
Assembly
 Components
Components


 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  ESRF
ESRF  / Beamline: BM14 / Wavelength: 0.97 Å
/ Beamline: BM14 / Wavelength: 0.97 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller












 PDBj
PDBj








