[English] 日本語
 Yorodumi
Yorodumi- PDB-1ohz: Cohesin-Dockerin complex from the cellulosome of Clostridium ther... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1ohz | ||||||
|---|---|---|---|---|---|---|---|
| Title | Cohesin-Dockerin complex from the cellulosome of Clostridium thermocellum | ||||||
 Components Components |
| ||||||
 Keywords Keywords | CELL ADHESION / COHESIN-DOCKERIN COMPLEX / COHESIN / DOCKERIN / CELLULOSOME / CLOSTRIDIUM THERMOCELLUM / CELLULOSE DEGRADATION | ||||||
| Function / homology |  Function and homology information Function and homology informationcellulosome / cellulose binding / endo-1,4-beta-xylanase activity / endo-1,4-beta-xylanase / xylan catabolic process / cellulose catabolic process / hydrolase activity, hydrolyzing O-glycosyl compounds / cell wall organization / extracellular region Similarity search - Function | ||||||
| Biological species |  CLOSTRIDIUM THERMOCELLUM (bacteria) CLOSTRIDIUM THERMOCELLUM (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.2 Å MOLECULAR REPLACEMENT / Resolution: 2.2 Å | ||||||
 Authors Authors | Carvalho, A.L. / Dias, F.M.V. / Prates, J.A.M. / Ferreira, L.M.A. / Gilbert, H.J. / Davies, G.J. / Romao, M.J. / Fontes, C.M.G.A. | ||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 2003 Journal: Proc.Natl.Acad.Sci.USA / Year: 2003Title: Cellulosome Assembly Revealed by the Crystal Structure of the Cohesin-Dockerin Complex Authors: Carvalho, A.L. / Dias, F.M.V. / Prates, J.A.M. / Ferreira, L.M.A. / Gilbert, H.J. / Davies, G.J. / Romao, M.J. / Fontes, C.M.G.A. | ||||||
| History |
|
- Structure visualization
Structure visualization
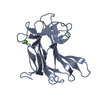
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1ohz.cif.gz 1ohz.cif.gz | 53.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1ohz.ent.gz pdb1ohz.ent.gz | 37.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1ohz.json.gz 1ohz.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/oh/1ohz https://data.pdbj.org/pub/pdb/validation_reports/oh/1ohz ftp://data.pdbj.org/pub/pdb/validation_reports/oh/1ohz ftp://data.pdbj.org/pub/pdb/validation_reports/oh/1ohz | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1anuS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| |||||||||
| Unit cell |
| |||||||||
| Components on special symmetry positions |
| |||||||||
| Details | THIS MAY BE THE RESULT OF CRYSTAL PACKING |
- Components
Components
-Protein , 2 types, 2 molecules AB
| #1: Protein | Mass: 17027.117 Da / Num. of mol.: 1 / Fragment: RESIDUES 181-340 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  CLOSTRIDIUM THERMOCELLUM (bacteria) / Production host: CLOSTRIDIUM THERMOCELLUM (bacteria) / Production host:  |
|---|---|
| #2: Protein | Mass: 6515.437 Da / Num. of mol.: 1 / Fragment: RESIDUES 733-791 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  CLOSTRIDIUM THERMOCELLUM (bacteria) / Production host: CLOSTRIDIUM THERMOCELLUM (bacteria) / Production host:  |
-Non-polymers , 5 types, 121 molecules 








| #3: Chemical | | #4: Chemical | ChemComp-EDO / | #5: Chemical | #6: Chemical | ChemComp-CL / | #7: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.32 Å3/Da / Density % sol: 62.97 % | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 7 / Details: pH 7.00 | ||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Method: unknown | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 110 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: BM30A / Wavelength: 0.976288 / Beamline: BM30A / Wavelength: 0.976288 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.976288 Å / Relative weight: 1 |
| Reflection | Resolution: 2.201→17.04 Å / Num. obs: 16170 / % possible obs: 99.8 % / Observed criterion σ(I): 2 / Redundancy: 11.9 % / Rmerge(I) obs: 0.072 / Net I/σ(I): 6.9 |
| Reflection shell | Resolution: 2.2→2.32 Å / Redundancy: 12 % / Rmerge(I) obs: 0.309 / Mean I/σ(I) obs: 1.9 / % possible all: 99.8 |
| Reflection | *PLUS Highest resolution: 2.2 Å / Lowest resolution: 17.04 Å / Redundancy: 11.9 % / Rmerge(I) obs: 0.072 |
| Reflection shell | *PLUS % possible obs: 99.8 % / Redundancy: 12 % / Rmerge(I) obs: 0.309 / Mean I/σ(I) obs: 1.9 |
- Processing
Processing
| Software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1ANU Resolution: 2.2→17 Å / Cross valid method: THROUGHOUT
| ||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.2→17 Å
| ||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 2.2 Å / Lowest resolution: 17 Å / % reflection Rfree: 5 % / Rfactor Rfree: 0.242 / Rfactor Rwork: 0.21 | ||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller
















 PDBj
PDBj





