| Entry | Database: PDB / ID: 1gkl
|
|---|
| Title | S954A mutant of the feruloyl esterase module from clostridium thermocellum complexed with ferulic acid |
|---|
 Components Components | ENDO-1,4-BETA-XYLANASE Y |
|---|
 Keywords Keywords | HYDROLASE / ESTERASE FAMILY 1 / INACTIVE MUTANT |
|---|
| Function / homology |  Function and homology information Function and homology information
Carbohydrate-binding, CenC-like / Carbohydrate binding domain / Esterase-like / Putative esterase / Clostridium cellulosome enzymes repeated domain signature. / Glycosyl hydrolases family 10, active site / Glycosyl hydrolases family 10 (GH10) active site. / Dockerin domain / Dockerin domain profile. / Dockerin type I domain ...Carbohydrate-binding, CenC-like / Carbohydrate binding domain / Esterase-like / Putative esterase / Clostridium cellulosome enzymes repeated domain signature. / Glycosyl hydrolases family 10, active site / Glycosyl hydrolases family 10 (GH10) active site. / Dockerin domain / Dockerin domain profile. / Dockerin type I domain / Dockerin type I repeat / Dockerin domain superfamily / Glycosyl hydrolases family 10 (GH10) domain profile. / Glycoside hydrolase family 10 / Glycoside hydrolase family 10 domain / Glycosyl hydrolase family 10 / Glycosyl hydrolase family 10 / Galactose-binding-like domain superfamily / Alpha/Beta hydrolase fold, catalytic domain / EF-Hand 1, calcium-binding site / EF-hand calcium-binding domain. / Alpha/Beta hydrolase fold / Glycoside hydrolase superfamily / Rossmann fold / 3-Layer(aba) Sandwich / Alpha BetaSimilarity search - Domain/homology |
|---|
| Biological species |  CLOSTRIDIUM THERMOCELLUM (bacteria) CLOSTRIDIUM THERMOCELLUM (bacteria) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.4 Å MOLECULAR REPLACEMENT / Resolution: 1.4 Å |
|---|
 Authors Authors | Prates, J.A.M. / Tarbouriech, N. / Charnock, S.J. / Fontes, C.M.G.A. / Ferreira, L.M.A. / Davies, G.J. |
|---|
 Citation Citation |  Journal: Structure / Year: 2001 Journal: Structure / Year: 2001
Title: The structure of the feruloyl esterase module of xylanase 10B from Clostridium thermocellum provides insights into substrate recognition.
Authors: Prates, J.A. / Tarbouriech, N. / Charnock, S.J. / Fontes, C.M. / Ferreira, L.M. / Davies, G.J. |
|---|
| History | | Deposition | Aug 15, 2001 | Deposition site: PDBE / Processing site: PDBE |
|---|
| Revision 1.0 | Dec 13, 2001 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Sep 7, 2011 | Group: Database references / Derived calculations ...Database references / Derived calculations / Non-polymer description / Other / Structure summary / Version format compliance |
|---|
| Revision 1.2 | Oct 9, 2019 | Group: Data collection / Database references / Other / Category: citation / citation_author / pdbx_database_status
Item: _citation.page_last / _citation.pdbx_database_id_DOI ..._citation.page_last / _citation.pdbx_database_id_DOI / _citation.title / _citation_author.name / _pdbx_database_status.status_code_sf |
|---|
| Revision 1.3 | May 8, 2024 | Group: Data collection / Database references / Derived calculations
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_struct_conn_angle / struct_conn / struct_site
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_struct_conn_angle.ptnr1_auth_asym_id / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr1_symmetry / _pdbx_struct_conn_angle.ptnr2_auth_asym_id / _pdbx_struct_conn_angle.ptnr2_auth_seq_id / _pdbx_struct_conn_angle.ptnr2_label_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.ptnr3_symmetry / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr1_symmetry / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id / _struct_conn.ptnr2_symmetry / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information CLOSTRIDIUM THERMOCELLUM (bacteria)
CLOSTRIDIUM THERMOCELLUM (bacteria) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.4 Å
MOLECULAR REPLACEMENT / Resolution: 1.4 Å  Authors
Authors Citation
Citation Journal: Structure / Year: 2001
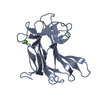
Journal: Structure / Year: 2001 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 1gkl.cif.gz
1gkl.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb1gkl.ent.gz
pdb1gkl.ent.gz PDB format
PDB format 1gkl.json.gz
1gkl.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/gk/1gkl
https://data.pdbj.org/pub/pdb/validation_reports/gk/1gkl ftp://data.pdbj.org/pub/pdb/validation_reports/gk/1gkl
ftp://data.pdbj.org/pub/pdb/validation_reports/gk/1gkl Links
Links Assembly
Assembly


 Components
Components CLOSTRIDIUM THERMOCELLUM (bacteria) / Strain: YS / Plasmid: PET-21A / Production host:
CLOSTRIDIUM THERMOCELLUM (bacteria) / Strain: YS / Plasmid: PET-21A / Production host: 








 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  SRS
SRS  / Beamline: PX14.2 / Wavelength: 0.978
/ Beamline: PX14.2 / Wavelength: 0.978  Processing
Processing MOLECULAR REPLACEMENT / Resolution: 1.4→19.92 Å / Cor.coef. Fo:Fc: 0.969 / Cor.coef. Fo:Fc free: 0.962 / SU B: 0.904 / SU ML: 0.019 / Cross valid method: THROUGHOUT / ESU R: 0.048 / ESU R Free: 0.044 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
MOLECULAR REPLACEMENT / Resolution: 1.4→19.92 Å / Cor.coef. Fo:Fc: 0.969 / Cor.coef. Fo:Fc free: 0.962 / SU B: 0.904 / SU ML: 0.019 / Cross valid method: THROUGHOUT / ESU R: 0.048 / ESU R Free: 0.044 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS Movie
Movie Controller
Controller















 PDBj
PDBj



