[English] 日本語
 Yorodumi
Yorodumi- PDB-5vf2: scFv 2D10 re-refined as a complex with trehalose replacing the or... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5vf2 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
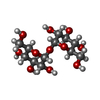
| Title | scFv 2D10 re-refined as a complex with trehalose replacing the original alpha-1,6-mannobiose | ||||||||||||
 Components Components | scFv 2D10 | ||||||||||||
 Keywords Keywords | IMMUNE SYSTEM / IMMUNE SYSTEM RE-REFINEMENT | ||||||||||||
| Function / homology | Immunoglobulins / Immunoglobulin-like / Sandwich / Mainly Beta / trehalose Function and homology information Function and homology information | ||||||||||||
| Biological species |  | ||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  FOURIER SYNTHESIS / Resolution: 1.55 Å FOURIER SYNTHESIS / Resolution: 1.55 Å | ||||||||||||
 Authors Authors | Porebski, P.J. / Wlodawer, A. / Dauter, Z. / Minor, W. / Stanfield, R. / Jaskolski, M. / Pozharski, E. / Weichenberger, C.X. / Rupp, B. | ||||||||||||
| Funding support |  United States, United States,  Austria, Austria,  Poland, 3items Poland, 3items
| ||||||||||||
 Citation Citation |  Journal: Chemistryselect / Year: 2016 Journal: Chemistryselect / Year: 2016Title: Antibodies Can Exploit Molecular Crowding to Bind New Antigens at Noncanonical Paratope Positions Authors: Vashisht, S. / Kumar, A. / Kaur, K.J. / Salunke, D.M. | ||||||||||||
| History |
| ||||||||||||
| Remark 0 | THIS ENTRY 5VF2 REFLECTS AN ALTERNATIVE MODELING OF THE ORIGINAL DATA IN 5I4F, DETERMINED BY S. ...THIS ENTRY 5VF2 REFLECTS AN ALTERNATIVE MODELING OF THE ORIGINAL DATA IN 5I4F, DETERMINED BY S.VASHISHT,A.KUMAR,K.J.KAUR,D.M.SALUNKE |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5vf2.cif.gz 5vf2.cif.gz | 120.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5vf2.ent.gz pdb5vf2.ent.gz | 91.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5vf2.json.gz 5vf2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/vf/5vf2 https://data.pdbj.org/pub/pdb/validation_reports/vf/5vf2 ftp://data.pdbj.org/pub/pdb/validation_reports/vf/5vf2 ftp://data.pdbj.org/pub/pdb/validation_reports/vf/5vf2 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5vehC  5vepC  5veqC  5verC  5vetC  5vf5C  5vgaC  5i4fS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| Unit cell |
| |||||||||
| Components on special symmetry positions |
|
- Components
Components
-Antibody / Sugars , 2 types, 3 molecules A
| #1: Antibody | Mass: 26894.973 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| #2: Polysaccharide |
-Non-polymers , 4 types, 323 molecules 






| #3: Chemical | ChemComp-MES / | ||||
|---|---|---|---|---|---|
| #4: Chemical | ChemComp-UNX / #5: Chemical | ChemComp-MG / | #6: Water | ChemComp-HOH / | |
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.72 Å3/Da / Density % sol: 54.72 % / Description: AUTHOR USED THE SF DATA FROM ENTRY 5I4F |
|---|---|
| Crystal grow | Temperature: 277.15 K / Method: vapor diffusion, hanging drop / pH: 6.5 Details: MES 6.5, Magnesium sulphate (1.5 M), potassium sodium tartrate tetrahydrate (0.1M). |
-Data collection
| Diffraction | Mean temperature: 173.15 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: BM14 / Wavelength: 0.97856 Å / Beamline: BM14 / Wavelength: 0.97856 Å |
| Detector | Type: MARMOSAIC 225 mm CCD / Detector: CCD / Date: Feb 20, 2014 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97856 Å / Relative weight: 1 |
| Reflection | Resolution: 1.549→50 Å / Num. obs: 41534 / % possible obs: 99.4 % / Redundancy: 5.5 % / Net I/σ(I): 39.67 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  FOURIER SYNTHESIS FOURIER SYNTHESISStarting model: 5I4F Resolution: 1.55→35.11 Å / Cor.coef. Fo:Fc: 0.974 / Cor.coef. Fo:Fc free: 0.965 / SU B: 2.486 / SU ML: 0.046 / Cross valid method: THROUGHOUT / ESU R: 0.07 / ESU R Free: 0.072 / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 26.12 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.55→35.11 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller










 PDBj
PDBj