[English] 日本語
 Yorodumi
Yorodumi- PDB-5ver: MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB S... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5ver | ||||||
|---|---|---|---|---|---|---|---|
| Title | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2Z | ||||||
 Components Components | Kynurenine--oxoglutarate transaminase 3 | ||||||
 Keywords Keywords | TRANSFERASE / AMINOTRANSFERASE III / MOUSE / RE-REFINEMENT | ||||||
| Function / homology |  Function and homology information Function and homology informationL-kynurenine catabolic process / kynurenine-glyoxylate transaminase / kynurenine-glyoxylate transaminase activity / cysteine-S-conjugate beta-lyase activity / cysteine-S-conjugate beta-lyase / kynurenine-oxoglutarate transaminase / kynurenine-oxoglutarate transaminase activity / kynurenine metabolic process / 2-oxoglutarate metabolic process / biosynthetic process ...L-kynurenine catabolic process / kynurenine-glyoxylate transaminase / kynurenine-glyoxylate transaminase activity / cysteine-S-conjugate beta-lyase activity / cysteine-S-conjugate beta-lyase / kynurenine-oxoglutarate transaminase / kynurenine-oxoglutarate transaminase activity / kynurenine metabolic process / 2-oxoglutarate metabolic process / biosynthetic process / amino acid metabolic process / pyridoxal phosphate binding / protein homodimerization activity / mitochondrion Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 2.81 Å SYNCHROTRON / Resolution: 2.81 Å | ||||||
 Authors Authors | Wlodawer, A. / Dauter, Z. / Minor, W. / Stanfield, R. / Porebski, P. / Jaskolski, M. / Pozharski, E. / Weichenberger, C.X. / Rupp, B. | ||||||
 Citation Citation | Journal: Mol. Cell. Biol. / Year: 2009 Title: Biochemical and structural properties of mouse kynurenine aminotransferase III. Authors: Han, Q. / Robinson, H. / Cai, T. / Tagle, D.A. / Li, J. | ||||||
| History |
| ||||||
| Remark 0 | This entry reflects an alternative modeling of the original data in 3E2Z, determined by Zhang, R., ...This entry reflects an alternative modeling of the original data in 3E2Z, determined by Zhang, R., Hatzos, C., Clancy, S., Joachimiak, A. | ||||||
| Remark 200 | SEE THE ORIGINAL DATA, ENTRY 3E2Z | ||||||
| Remark 280 | SEE THE ORIGINAL DATA, ENTRY 3E2Z |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5ver.cif.gz 5ver.cif.gz | 174.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5ver.ent.gz pdb5ver.ent.gz | 142.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5ver.json.gz 5ver.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  5ver_validation.pdf.gz 5ver_validation.pdf.gz | 506 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  5ver_full_validation.pdf.gz 5ver_full_validation.pdf.gz | 522.8 KB | Display | |
| Data in XML |  5ver_validation.xml.gz 5ver_validation.xml.gz | 33.3 KB | Display | |
| Data in CIF |  5ver_validation.cif.gz 5ver_validation.cif.gz | 45.4 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ve/5ver https://data.pdbj.org/pub/pdb/validation_reports/ve/5ver ftp://data.pdbj.org/pub/pdb/validation_reports/ve/5ver ftp://data.pdbj.org/pub/pdb/validation_reports/ve/5ver | HTTPS FTP |
-Related structure data
| Related structure data |  5vehC  5vepC  5veqC  5vetC  5vf2C  5vf5C  5vgaC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 46224.977 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   References: UniProt: Q71RI9, kynurenine-oxoglutarate transaminase, cysteine-S-conjugate beta-lyase, kynurenine-glyoxylate transaminase |
|---|
-Non-polymers , 8 types, 115 molecules 




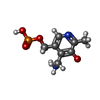









| #2: Chemical | ChemComp-PLP / | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| #3: Chemical | | #4: Chemical | ChemComp-PGE / | #5: Chemical | ChemComp-GOL / #6: Chemical | ChemComp-CA / #7: Chemical | ChemComp-PMP / | #8: Chemical | ChemComp-PEG / | #9: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.63 Å3/Da / Density % sol: 53.23 % |
|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop / pH: 7.5 Details: 21% PEG 400, 150 MM CACL2, 10%, GLYCEROL, 100 MM HEPES, PH 7.5 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  NSLS NSLS  / Beamline: X29A / Wavelength: 1.0809 Å / Beamline: X29A / Wavelength: 1.0809 Å |
| Detector | Type: ADSC QUANTUM 315 / Detector: CCD / Date: Jun 27, 2008 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.0809 Å / Relative weight: 1 |
| Reflection | Resolution: 2.8→29.64 Å / Num. obs: 22779 / % possible obs: 95.6 % / Redundancy: 11.9 % / Net I/σ(I): 11.2 |
- Processing
Processing
| Software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 2.81→29.64 Å / SU B: 13.552 / SU ML: 0.264 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R Free: 0.396
| ||||||||||||||||||||
| Displacement parameters | Biso mean: 30.46 Å2
| ||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.81→29.64 Å
|
 Movie
Movie Controller
Controller













 PDBj
PDBj






