+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5vc9 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Zinc finger of human CXXC4 in complex with CpG DNA | ||||||
 Components Components |
| ||||||
 Keywords Keywords | DNA BINDING PROTEIN / CXXC / DNA / Structural Genomics / Structural Genomics Consortium / SGC | ||||||
| Function / homology |  Function and homology information Function and homology informationzygotic specification of dorsal/ventral axis / Negative regulation of TCF-dependent signaling by DVL-interacting proteins / methyl-CpG binding / negative regulation of Wnt signaling pathway / PDZ domain binding / Wnt signaling pathway / cytoplasmic vesicle / zinc ion binding / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human)synthetic construct (others) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.1 Å MOLECULAR REPLACEMENT / Resolution: 2.1 Å | ||||||
 Authors Authors | Liu, K. / Xu, C. / Tempel, W. / Walker, J.R. / Arrowsmith, C.H. / Bountra, C. / Edwards, A.M. / Min, J. / Structural Genomics Consortium (SGC) | ||||||
 Citation Citation |  Journal: Structure / Year: 2018 Journal: Structure / Year: 2018Title: DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants. Authors: Xu, C. / Liu, K. / Lei, M. / Yang, A. / Li, Y. / Hughes, T.R. / Min, J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5vc9.cif.gz 5vc9.cif.gz | 101.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5vc9.ent.gz pdb5vc9.ent.gz | 74.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5vc9.json.gz 5vc9.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  5vc9_validation.pdf.gz 5vc9_validation.pdf.gz | 442.2 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  5vc9_full_validation.pdf.gz 5vc9_full_validation.pdf.gz | 442.2 KB | Display | |
| Data in XML |  5vc9_validation.xml.gz 5vc9_validation.xml.gz | 7.1 KB | Display | |
| Data in CIF |  5vc9_validation.cif.gz 5vc9_validation.cif.gz | 9.4 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/vc/5vc9 https://data.pdbj.org/pub/pdb/validation_reports/vc/5vc9 ftp://data.pdbj.org/pub/pdb/validation_reports/vc/5vc9 ftp://data.pdbj.org/pub/pdb/validation_reports/vc/5vc9 | HTTPS FTP |
-Related structure data
| Related structure data |  4nw3C  4o64C  4pziC  4z3cC  5w9qC  5w9sC  6asbC  6asdC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 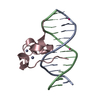
| ||||||||
| 2 | 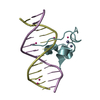
| ||||||||
| Unit cell |
|
- Components
Components
| #1: DNA chain | Mass: 3663.392 Da / Num. of mol.: 4 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) #2: Protein/peptide | Mass: 5611.951 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: CXXC4, IDAX / Plasmid: pET28-MHL / Production host: Homo sapiens (human) / Gene: CXXC4, IDAX / Plasmid: pET28-MHL / Production host:  #3: Chemical | ChemComp-UNX / #4: Chemical | ChemComp-ZN / #5: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.4 Å3/Da / Density % sol: 49.3 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion / pH: 7.5 Details: 30% PEG-1500, 5% MPD, 0.2 M sodium chloride, 0.1 M HEPES |
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 19-ID / Wavelength: 0.97918 Å / Beamline: 19-ID / Wavelength: 0.97918 Å | ||||||||||||||||||||||||||||||
| Detector | Type: ADSC QUANTUM 315 / Detector: CCD / Date: Jun 19, 2013 | ||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 0.97918 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||
| Reflection | Resolution: 2.1→27.24 Å / Num. obs: 13389 / % possible obs: 97.2 % / Redundancy: 2.1 % / CC1/2: 0.995 / Rmerge(I) obs: 0.061 / Rpim(I) all: 0.054 / Rrim(I) all: 0.081 / Net I/σ(I): 10.1 / Num. measured all: 28017 / Scaling rejects: 0 | ||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: unpublished model of related protein-DNA complex Resolution: 2.1→27.2 Å / Cor.coef. Fo:Fc: 0.957 / Cor.coef. Fo:Fc free: 0.937 / SU B: 13.841 / SU ML: 0.174 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.245 / ESU R Free: 0.196 Details: coot was used for interactive model building. Model geometry was assessed with phenix.molprobity.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 97.52 Å2 / Biso mean: 45.818 Å2 / Biso min: 14.09 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.1→27.2 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.1→2.154 Å / Total num. of bins used: 20
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj











































