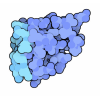
Entry Database : PDB / ID : 5juhTitle Crystal structure of C-terminal domain (RV) of MpAFP Antifreeze protein Keywords / / / Function / homology Function Domain/homology Component
/ / / / / / / Biological species Marinomonas primoryensis (bacteria)Method / / Resolution : 1.35 Å Authors Guo, S. Funding support Organization Grant number Country Canadian Institutes of Health Research (CIHR)
Journal : Sci Adv / Year : 2017Title : Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.Authors: Guo, S. / Stevens, C.A. / Vance, T.D.R. / Olijve, L.L.C. / Graham, L.A. / Campbell, R.L. / Yazdi, S.R. / Escobedo, C. / Bar-Dolev, M. / Yashunsky, V. / Braslavsky, I. / Langelaan, D.N. / ... Authors : Guo, S. / Stevens, C.A. / Vance, T.D.R. / Olijve, L.L.C. / Graham, L.A. / Campbell, R.L. / Yazdi, S.R. / Escobedo, C. / Bar-Dolev, M. / Yashunsky, V. / Braslavsky, I. / Langelaan, D.N. / Smith, S.P. / Allingham, J.S. / Voets, I.K. / Davies, P.L. History Deposition May 10, 2016 Deposition site / Processing site Revision 1.0 Jul 19, 2017 Provider / Type Revision 1.1 Sep 6, 2017 Group / Category / citation_authorItem _citation.country / _citation.journal_abbrev ... _citation.country / _citation.journal_abbrev / _citation.journal_id_CSD / _citation.journal_id_ISSN / _citation.journal_volume / _citation.page_first / _citation.page_last / _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation.year Revision 1.2 Sep 20, 2017 Group / Category / Item Revision 1.3 Jan 8, 2020 Group / Category / Item Revision 1.4 Mar 6, 2024 Group / Database references / Derived calculationsCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_struct_conn_angle / struct_conn Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_alt_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr1_symmetry / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_alt_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.ptnr3_symmetry / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.pdbx_ptnr2_label_alt_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_symmetry
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Marinomonas primoryensis (bacteria)
Marinomonas primoryensis (bacteria) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 1.35 Å
SYNCHROTRON / Resolution: 1.35 Å  Authors
Authors Canada, 1items
Canada, 1items  Citation
Citation Journal: Sci Adv / Year: 2017
Journal: Sci Adv / Year: 2017 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5juh.cif.gz
5juh.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5juh.ent.gz
pdb5juh.ent.gz PDB format
PDB format 5juh.json.gz
5juh.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/ju/5juh
https://data.pdbj.org/pub/pdb/validation_reports/ju/5juh ftp://data.pdbj.org/pub/pdb/validation_reports/ju/5juh
ftp://data.pdbj.org/pub/pdb/validation_reports/ju/5juh Links
Links Assembly
Assembly
 Components
Components Marinomonas primoryensis (bacteria) / Production host:
Marinomonas primoryensis (bacteria) / Production host: 
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  CLSI
CLSI  / Beamline: 08ID-1 / Wavelength: 0.97949 Å
/ Beamline: 08ID-1 / Wavelength: 0.97949 Å Processing
Processing Movie
Movie Controller
Controller
















 PDBj
PDBj