| Entry | Database: PDB / ID: 5d3d
|
|---|
| Title | Crystal structure of Staphylococcal Superantigen-Like protein 3 |
|---|
 Components Components | Staphylococcal Superantigen-Like protein 3 |
|---|
 Keywords Keywords | IMMUNE SYSTEM / Superantigens / Superantigen-like proteins / SSL / SSL3 / Toll-Like Receptor 2 / TLR2 / Immunology / Inflammation / Inhibition |
|---|
| Function / homology |  Function and homology information Function and homology information
Staphylococcus aureus exotoxin / Staphylococcal superantigen-like OB-fold domain / Staphylococcal superantigen-like OB-fold domain / Ubiquitin-like (UB roll) - #120 / Staphylococcal enterotoxin/Streptococcal pyrogenic exotoxin, conserved site / Staphyloccocal enterotoxin/Streptococcal pyrogenic exotoxin signature 2. / Superantigen, staphylococcal/streptococcal toxin, bacterial / Staphylococcal/Streptococcal toxin, beta-grasp domain / Staphylococcal/Streptococcal toxin, beta-grasp domain / Superantigen toxin, C-terminal ...Staphylococcus aureus exotoxin / Staphylococcal superantigen-like OB-fold domain / Staphylococcal superantigen-like OB-fold domain / Ubiquitin-like (UB roll) - #120 / Staphylococcal enterotoxin/Streptococcal pyrogenic exotoxin, conserved site / Staphyloccocal enterotoxin/Streptococcal pyrogenic exotoxin signature 2. / Superantigen, staphylococcal/streptococcal toxin, bacterial / Staphylococcal/Streptococcal toxin, beta-grasp domain / Staphylococcal/Streptococcal toxin, beta-grasp domain / Superantigen toxin, C-terminal / OB fold (Dihydrolipoamide Acetyltransferase, E2P) - #110 / Enterotoxin / Ubiquitin-like (UB roll) / OB fold (Dihydrolipoamide Acetyltransferase, E2P) / Roll / Beta Barrel / Mainly Beta / Alpha BetaSimilarity search - Domain/homology |
|---|
| Biological species |   Staphylococcus aureus (bacteria) Staphylococcus aureus (bacteria) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.94 Å MOLECULAR REPLACEMENT / Resolution: 1.94 Å |
|---|
 Authors Authors | Feitsma, L.J. / Huizinga, E.G. |
|---|
| Funding support |  Netherlands, 3items Netherlands, 3items | Organization | Grant number | Country |
|---|
| Netherlands Organization for Scientific Research | ECHO Grant 700.58.006 |  Netherlands Netherlands | | Netherlands Organization for Health Research and Development | ZonMw Grant 205200004 |  Netherlands Netherlands | | Dutch Top Institute Pharma Project | D1-101 |  Netherlands Netherlands |
|
|---|
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 2015 Journal: Proc.Natl.Acad.Sci.USA / Year: 2015
Title: Structural basis for inhibition of TLR2 by staphylococcal superantigen-like protein 3 (SSL3).
Authors: Koymans, K.J. / Feitsma, L.J. / Brondijk, T.H. / Aerts, P.C. / Lukkien, E. / Lossl, P. / van Kessel, K.P. / de Haas, C.J. / van Strijp, J.A. / Huizinga, E.G. |
|---|
| History | | Deposition | Aug 6, 2015 | Deposition site: RCSB / Processing site: PDBE |
|---|
| Revision 1.0 | Aug 19, 2015 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Sep 2, 2015 | Group: Database references |
|---|
| Revision 1.2 | Sep 9, 2015 | Group: Database references |
|---|
| Revision 1.3 | May 1, 2024 | Group: Data collection / Database references / Refinement description
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession |
|---|
|
|---|
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.94 Å
MOLECULAR REPLACEMENT / Resolution: 1.94 Å  Authors
Authors Netherlands, 3items
Netherlands, 3items  Citation
Citation Journal: Proc.Natl.Acad.Sci.USA / Year: 2015
Journal: Proc.Natl.Acad.Sci.USA / Year: 2015 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5d3d.cif.gz
5d3d.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5d3d.ent.gz
pdb5d3d.ent.gz PDB format
PDB format 5d3d.json.gz
5d3d.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/d3/5d3d
https://data.pdbj.org/pub/pdb/validation_reports/d3/5d3d ftp://data.pdbj.org/pub/pdb/validation_reports/d3/5d3d
ftp://data.pdbj.org/pub/pdb/validation_reports/d3/5d3d Links
Links Assembly
Assembly
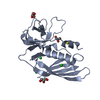

 Components
Components Staphylococcus aureus (strain NCTC 8325) (bacteria)
Staphylococcus aureus (strain NCTC 8325) (bacteria)









 X-RAY DIFFRACTION
X-RAY DIFFRACTION Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  SLS
SLS  / Beamline: X06SA / Wavelength: 0.9999 Å
/ Beamline: X06SA / Wavelength: 0.9999 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller







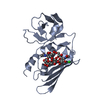






 PDBj
PDBj




