[English] 日本語
 Yorodumi
Yorodumi- PDB-5b8i: Crystal structure of Calcineurin A and Calcineurin B in complex w... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5b8i | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of Calcineurin A and Calcineurin B in complex with FKBP12 and FK506 from Coccidioides immitis RS | ||||||
 Components Components |
| ||||||
 Keywords Keywords | HYDROLASE / SSGCID / NIH / NIAID / SBRI / UW / BERYLLIUM / PHOSPHATASE / CALCINEURIN / FKBP12 / FK506 / Structural Genomics / Seattle Structural Genomics Center for Infectious Disease | ||||||
| Function / homology |  Function and homology information Function and homology informationhistone H2AXS139 phosphatase activity / RNA polymerase II CTD heptapeptide repeat Y1 phosphatase activity / RNA polymerase II CTD heptapeptide repeat T4 phosphatase activity / RNA polymerase II CTD heptapeptide repeat P3 isomerase activity / RNA polymerase II CTD heptapeptide repeat P6 isomerase activity / MAP kinase serine/threonine phosphatase activity / myosin phosphatase activity / calmodulin-dependent protein phosphatase activity / RNA polymerase II CTD heptapeptide repeat S2 phosphatase activity / RNA polymerase II CTD heptapeptide repeat S7 phosphatase activity ...histone H2AXS139 phosphatase activity / RNA polymerase II CTD heptapeptide repeat Y1 phosphatase activity / RNA polymerase II CTD heptapeptide repeat T4 phosphatase activity / RNA polymerase II CTD heptapeptide repeat P3 isomerase activity / RNA polymerase II CTD heptapeptide repeat P6 isomerase activity / MAP kinase serine/threonine phosphatase activity / myosin phosphatase activity / calmodulin-dependent protein phosphatase activity / RNA polymerase II CTD heptapeptide repeat S2 phosphatase activity / RNA polymerase II CTD heptapeptide repeat S7 phosphatase activity / RNA polymerase II CTD heptapeptide repeat S5 phosphatase activity / protein-serine/threonine phosphatase / calcineurin-mediated signaling / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / peptidylprolyl isomerase / small-subunit processome / calmodulin binding / calcium ion binding / nucleolus / metal ion binding / cytoplasm Similarity search - Function | ||||||
| Biological species |  Coccidioides immitis (fungus) Coccidioides immitis (fungus) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.85 Å MOLECULAR REPLACEMENT / Resolution: 1.85 Å | ||||||
 Authors Authors | Seattle Structural Genomics Center for Infectious Disease (SSGCID) / Fox III, D. / Dranow, D.M. / Lorimer, D.D. / Edwards, T.E. | ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Harnessing calcineurin-FK506-FKBP12 crystal structures from invasive fungal pathogens to develop antifungal agents. Authors: Juvvadi, P.R. / Fox 3rd, D. / Bobay, B.G. / Hoy, M.J. / Gobeil, S.M.C. / Venters, R.A. / Chang, Z. / Lin, J.J. / Averette, A.F. / Cole, D.C. / Barrington, B.C. / Wheaton, J.D. / Ciofani, M. ...Authors: Juvvadi, P.R. / Fox 3rd, D. / Bobay, B.G. / Hoy, M.J. / Gobeil, S.M.C. / Venters, R.A. / Chang, Z. / Lin, J.J. / Averette, A.F. / Cole, D.C. / Barrington, B.C. / Wheaton, J.D. / Ciofani, M. / Trzoss, M. / Li, X. / Lee, S.C. / Chen, Y.L. / Mutz, M. / Spicer, L.D. / Schumacher, M.A. / Heitman, J. / Steinbach, W.J. #1:  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Harnessing calcineurin-FK506-FKBP12 crystal structures from invasive fungal pathogens to develop antifungal agents Authors: Juvvadi, P.R. / Fox III, D. / Bobay, B.G. / Hoy, M.J. / Gobeil, S.M. / Venters, R.A. / Chang, Z. / Lin, J.J. / Averette, A.F. / Cole, D.C. / Barrington, B.C. / Wheaton, J.D. / Ciofani, M. / ...Authors: Juvvadi, P.R. / Fox III, D. / Bobay, B.G. / Hoy, M.J. / Gobeil, S.M. / Venters, R.A. / Chang, Z. / Lin, J.J. / Averette, A.F. / Cole, D.C. / Barrington, B.C. / Wheaton, J.D. / Ciofani, M. / Trzoss, M. / Li, X. / Lee, S.C. / Chen, Y. / Mutz, M. / Spicer, L.D. / Schumacher, M.A. / Heitman, J. / Steinbach, W.J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5b8i.cif.gz 5b8i.cif.gz | 301 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5b8i.ent.gz pdb5b8i.ent.gz | 239.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5b8i.json.gz 5b8i.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/b8/5b8i https://data.pdbj.org/pub/pdb/validation_reports/b8/5b8i ftp://data.pdbj.org/pub/pdb/validation_reports/b8/5b8i ftp://data.pdbj.org/pub/pdb/validation_reports/b8/5b8i | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6tz6C  6tz7C  6tz8C  1tcoS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data | |
| Other databases |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Details | Size exclusion chromatography indicates trimer |
- Components
Components
-Protein , 3 types, 3 molecules ABC
| #1: Protein | Mass: 45097.133 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Coccidioides immitis (strain RS) (fungus) Coccidioides immitis (strain RS) (fungus)Strain: RS / Gene: CIMG_06667 / Plasmid: pEMB36 / Production host:  Trichoplusia ni (cabbage looper) / References: UniProt: J3K9C5, UniProt: J3K8M7*PLUS Trichoplusia ni (cabbage looper) / References: UniProt: J3K9C5, UniProt: J3K8M7*PLUS |
|---|---|
| #2: Protein | Mass: 19502.895 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Coccidioides immitis (strain RS) (fungus) Coccidioides immitis (strain RS) (fungus)Strain: RS / Gene: CIMG_02704 / Plasmid: pEMB36 / Production host:  Trichoplusia ni (cabbage looper) / References: UniProt: A0A0D8JSK0 Trichoplusia ni (cabbage looper) / References: UniProt: A0A0D8JSK0 |
| #3: Protein | Mass: 14458.537 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Coccidioides immitis (strain RS) (fungus) Coccidioides immitis (strain RS) (fungus)Strain: RS / Gene: CIMG_08666 / Production host:  Trichoplusia ni (cabbage looper) / References: UniProt: J3K5Z5, peptidylprolyl isomerase Trichoplusia ni (cabbage looper) / References: UniProt: J3K5Z5, peptidylprolyl isomerase |
-Non-polymers , 7 types, 707 molecules 




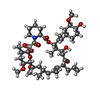







| #4: Chemical | ChemComp-ZN / | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| #5: Chemical | ChemComp-FE / | ||||||||
| #6: Chemical | | #7: Chemical | ChemComp-EDO / #8: Chemical | ChemComp-CA / #9: Chemical | ChemComp-FK5 / | #10: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3 Å3/Da / Density % sol: 58.96 % |
|---|---|
| Crystal grow | Temperature: 289 K / Method: vapor diffusion, sitting drop / pH: 6.5 Details: COCCIDIOIDES IMMITIS, COMPLEX NAMED COIMA.00174.A.GM11, AND COMPOSED OF CALCINEURIN A (COIMA.00174.A.GM11), CALCINEURIN B (COIMA.01011.A.GJ11), AND FKBP12 (COIMA.18272.A.GK11) PURIFIED IN ...Details: COCCIDIOIDES IMMITIS, COMPLEX NAMED COIMA.00174.A.GM11, AND COMPOSED OF CALCINEURIN A (COIMA.00174.A.GM11), CALCINEURIN B (COIMA.01011.A.GJ11), AND FKBP12 (COIMA.18272.A.GK11) PURIFIED IN THE PRESENCE OF FK506/ TACROLIMUS (PROTEIN BATCH #D00367) AT 10.81 MG/ML. PROTEIN BUFFER INCLUDES 25MM HEPES PH 8.0, 50 MM NACL, 5.0MM CACL2, 0.5MM TCEP, AND 5UM FK506. THE PROTEIN COMPLEX WAS CRYSTALLIZED AGAINST THE SPARSE MATRIX SCREEN PROPLEX V/V PROPANOL-1 AND CRYO-PROTECTED WITH 20% ETHYLENE GLYCOL; CRYSTAL TRACKING ID 261098D2 (HLC3-10), VAPOR DIFFUSION, SITTING DROP, TEMPERATURE 289K PH range: 6.5 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 21-ID-G / Wavelength: 0.97856 Å / Beamline: 21-ID-G / Wavelength: 0.97856 Å |
| Detector | Type: MARMOSAIC 300 mm CCD / Detector: CCD / Date: Feb 25, 2015 |
| Radiation | Monochromator: Diamond [111] / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97856 Å / Relative weight: 1 |
| Reflection | Resolution: 1.85→50 Å / Num. obs: 81827 / % possible obs: 100 % / Observed criterion σ(I): -3 / Redundancy: 7.39 % / Biso Wilson estimate: 26.54 Å2 / Rmerge(I) obs: 0.087 / Net I/σ(I): 15.7 |
| Reflection shell | Resolution: 1.85→1.9 Å / Rmerge(I) obs: 0.479 / Mean I/σ(I) obs: 4.2 / % possible all: 100 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1TCO Resolution: 1.85→45.27 Å / SU ML: 0.15 / Cross valid method: FREE R-VALUE / Phase error: 15.66 / Stereochemistry target values: ML
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 27.08 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.85→45.27 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.85→1.8718 Å / Phase error: 16.55
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj







